7N27
 
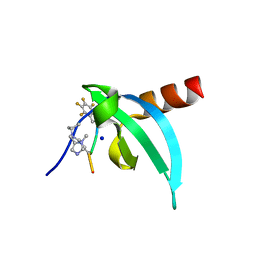 | | Crystal Structure of chromodomain of CDYL in complex with inhibitor UNC6261 | | Descriptor: | Isoform 2 of Chromodomain Y-like protein, SODIUM ION, UNKNOWN ATOM OR ION, ... | | Authors: | Beldar, S, Dong, A, Loppnau, P, Min, J, Arrowsmith, C.H, Edwards, A.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of chromodomain of CDYL in complex with inhibitor UNC6261
To Be Published
|
|
5AMF
 
 | | Crystal structure of the bromodomain of human surface epitope engineered BRD1A in complex with 3D Consortium fragment Ethyl 4,5,6,7- tetrahydro-1H-indazole-5-carboxylate (SGC - Diamond I04-1 fragment screening) | | Descriptor: | BROMODOMAIN-CONTAINING PROTEIN 1, ETHYL (5R)-4,5,6,7-TETRAHYDRO-1H-INDAZOLE-5-CARBOXYLATE, SODIUM ION | | Authors: | Pearce, N.M, Fairhead, M, Strain-Damerell, C, Talon, R, Wright, N, Ng, J.T, Bradley, A, Cox, O, Bowkett, D, Collins, P, Brandao-Neto, J, Douangamath, A, Krojer, T, Burgess-Brown, N, Brennan, P, Arrowsmith, C.H, Edwards, E, Bountra, C, von Delft, F. | | Deposit date: | 2015-03-10 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the Bromodomain of Human Surface Epitope Engineered Brd1A in Complex with 3D Consortium Fragment Ethyl 4,5,6,7-Tetrahydro-1H-Indazole-5-Carboxylate
To be Published
|
|
5AME
 
 | | Crystal structure of the bromodomain of human surface epitope engineered BRD1A in complex with 3D Consortium fragment 4-acetyl- piperazin-2-one (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 4-acetyl-piperazin-2-one, BROMODOMAIN-CONTAINING PROTEIN 1, SODIUM ION | | Authors: | Pearce, N.M, Fairhead, M, Strain-Damerell, C, Talon, R, Wright, N, Ng, J.T, Bradley, A, Cox, O, Bowkett, D, Collins, P, Brandao-Neto, J, Douangamath, A, Krojer, T, Burgess-Brown, N, Brennan, P, Arrowsmith, C.H, Edwards, E, Bountra, C, von Delft, F. | | Deposit date: | 2015-03-10 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.578 Å) | | Cite: | Crystal Structure of the Bromodomain of Human Surface Epitope Engineered Brd1A in Complex with 3D Consortium Fragment 4-Acetyl-Piperazin-2-One
To be Published
|
|
4BCE
 
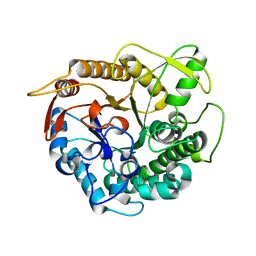 | | crystal structure of Ttb-gly N282T mutant | | Descriptor: | BETA-GLUCOSIDASE | | Authors: | Teze, D, Tran, V, Tellier, C, Dion, M, Leroux, C, Roncza, J, Czjzek, M. | | Deposit date: | 2012-10-02 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Semi-Rational Approach for Converting a Gh1 Beta-Glycosidase Into a Beta-Transglycosidase.
Protein Eng.Des.Sel., 27, 2014
|
|
5AMQ
 
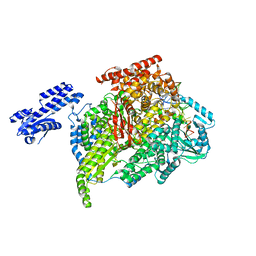 | |
1R67
 
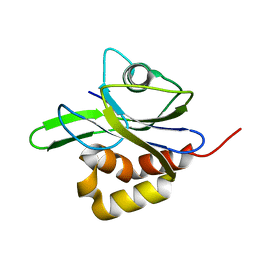 | | Y104A MUTANT OF E.COLI IPP ISOMERASE | | Descriptor: | Isopentenyl-diphosphate delta-isomerase, MAGNESIUM ION | | Authors: | Wouters, J. | | Deposit date: | 2003-10-15 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural role for Tyr-104 in Escherichia coli isopentenyl-diphosphate isomerase: site-directed mutagenesis, enzymology, and protein crystallography.
J.Biol.Chem., 281, 2006
|
|
7MYY
 
 | | Crystal Structure of HIV-1 PRS17 with GRL-142 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Agniswamy, J, Weber, I.T. | | Deposit date: | 2021-05-22 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Novel HIV PR inhibitors with C4-substituted bis-THF and bis-fluoro-benzyl target the two active site mutations of highly drug resistant mutant PR S17 .
Biochem.Biophys.Res.Commun., 566, 2021
|
|
3U13
 
 | | Crystal Structure of de Novo design of cystein esterase ECH13 at the resolution 1.6A, Northeast Structural Genomics Consortium Target OR51 | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Sahdev, S, Xiao, R, Kohan, E, Richter, F, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-29 | | Release date: | 2011-11-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Computational design of catalytic dyads and oxyanion holes for ester hydrolysis.
J.Am.Chem.Soc., 134, 2012
|
|
2O8J
 
 | | Human euchromatic histone methyltransferase 2 | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Min, J, Wu, H, Antoshenko, T, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-12-12 | | Release date: | 2007-01-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural biology of human H3K9 methyltransferases
Plos One, 5, 2010
|
|
7N98
 
 | | Cryo-EM structure of MFSD2A | | Descriptor: | Sodium-dependent lysophosphatidylcholine symporter 1 | | Authors: | Zhang, J, Feng, L. | | Deposit date: | 2021-06-17 | | Release date: | 2021-08-04 | | Last modified: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure and mechanism of blood-brain-barrier lipid transporter MFSD2A.
Nature, 596, 2021
|
|
7MYP
 
 | | Crystal Structure of HIV-1 PRS17 with GRL-44-10A | | Descriptor: | (3R,3aS,4R,6aR)-4-methoxyhexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Agniswamy, J, Weber, I.T. | | Deposit date: | 2021-05-21 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Novel HIV PR inhibitors with C4-substituted bis-THF and bis-fluoro-benzyl target the two active site mutations of highly drug resistant mutant PR S17 .
Biochem.Biophys.Res.Commun., 566, 2021
|
|
4TW6
 
 | | The Fk1 domain of FKBP51 in complex with iFit1 | | Descriptor: | (3-{(1R)-3-(3,4-dimethoxyphenyl)-1-[({(2S)-1-[(2S)-2-(3,4,5-trimethoxyphenyl)pent-4-enoyl]piperidin-2-yl}carbonyl)oxy]propyl}phenoxy)acetic acid, GLYCEROL, Peptidyl-prolyl cis-trans isomerase FKBP5, ... | | Authors: | Gaali, S, Kirschner, A, Cuboni, S, Hartmann, J, Kozany, C, Balsevich, G, Namendorf, C, Fernandez-Vizarra, P, Almeida, O.F.X, Ruehter, G, Uhr, M, Schmidt, M.V, Touma, C, Bracher, A, Hausch, F. | | Deposit date: | 2014-06-30 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Selective inhibitors of the FK506-binding protein 51 by induced fit.
Nat.Chem.Biol., 11, 2015
|
|
5DZD
 
 | | Crystal Structure of WW4 domain of ITCH in complex with TXNIP peptide | | Descriptor: | E3 ubiquitin-protein ligase Itchy homolog, Thioredoxin-interacting protein, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Dong, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-25 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal Structure of WW4 domain of ITCH in complex with TXNIP peptide
To be Published
|
|
2LEA
 
 | | Solution structure of human SRSF2 (SC35) RRM | | Descriptor: | Serine/arginine-rich splicing factor 2 | | Authors: | Daubner, G.M, Clery, A, Jayne, S, Stevenin, J, Allain, F.H.-T. | | Deposit date: | 2011-06-15 | | Release date: | 2011-11-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A syn-anti conformational difference allows SRSF2 to recognize guanines and cytosines equally well.
Embo J., 31, 2012
|
|
4BED
 
 | |
4TWO
 
 | | Human EphA3 Kinase domain in complex with compound 164 | | Descriptor: | 5-{[3-carbamoyl-4-(3,4-dimethylphenyl)-5-methylthiophen-2-yl]amino}-5-oxopentanoic acid, Ephrin type-A receptor 3 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2014-07-01 | | Release date: | 2015-05-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Structural Analysis of the Binding of Type I, I1/2, and II Inhibitors to Eph Tyrosine Kinases.
Acs Med.Chem.Lett., 6, 2015
|
|
5E1B
 
 | | Crystal structure of NRMT1 in complex with SPKRIA peptide | | Descriptor: | GLYCEROL, N-terminal Xaa-Pro-Lys N-methyltransferase 1, RCC1, ... | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-29 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for substrate recognition by the human N-terminal methyltransferase 1.
Genes Dev., 29, 2015
|
|
5E1P
 
 | | Ca(2+)-Calmodulin from Paramecium tetraurelia qFit disorder model | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Calmodulin | | Authors: | Lin, J, van den Bedem, H, Brunger, A.T, Wilson, M.A. | | Deposit date: | 2015-09-29 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Atomic resolution experimental phase information reveals extensive disorder and bound 2-methyl-2,4-pentanediol in Ca(2+)-calmodulin.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
1RPY
 
 | | CRYSTAL STRUCTURE OF THE DIMERIC SH2 DOMAIN OF APS | | Descriptor: | SULFATE ION, adaptor protein APS | | Authors: | Hu, J, Liu, J, Ghirlando, R, Saltiel, A.R, Hubbard, S.R. | | Deposit date: | 2003-12-03 | | Release date: | 2003-12-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for recruitment of the adaptor protein APS to the activated insulin receptor.
Mol.Cell, 12, 2003
|
|
7NJ0
 
 | | CryoEM structure of the human Separase-Cdk1-cyclin B1-Cks1 complex | | Descriptor: | Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 1, G2/mitotic-specific cyclin-B1,G2/mitotic-specific cyclin-B1, ... | | Authors: | Yu, J, Raia, P, Ghent, C.M, Raisch, T, Sadian, Y, Barford, D, Raunser, S, Morgan, D.O, Boland, A. | | Deposit date: | 2021-02-14 | | Release date: | 2021-08-04 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of human separase regulation by securin and CDK1-cyclin B1.
Nature, 596, 2021
|
|
4TZ9
 
 | | Structure of Metallo-beta-lactamase | | Descriptor: | CADMIUM ION, COBALT (II) ION, Class B metallo-beta-lactamase, ... | | Authors: | Ferguson, J.A, Brem, J, Makena, A, McDonough, A.M, Schofield, C.J. | | Deposit date: | 2014-07-09 | | Release date: | 2015-07-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1269 Å) | | Cite: | Structure of Metallo-beta-lactamase
To Be Published
|
|
4TZE
 
 | | Structure of metallo-beta-lactamase | | Descriptor: | Class B carbapenemase NDM-5, ZINC ION | | Authors: | Ferguson, J.A, Makena, A, Brem, J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2014-07-10 | | Release date: | 2015-07-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.574 Å) | | Cite: | stucuture of metallo-beta-lactamase
To Be Published
|
|
5E5W
 
 | | Hemagglutinin-esterase-fusion mutant structure of influenza D virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin-esterase, ... | | Authors: | Song, H, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2015-10-09 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An Open Receptor-Binding Cavity of Hemagglutinin-Esterase-Fusion Glycoprotein from Newly-Identified Influenza D Virus: Basis for Its Broad Cell Tropism
PLoS Pathog., 12, 2016
|
|
7NJ1
 
 | | CryoEM structure of the human Separase-Securin complex | | Descriptor: | Securin, Separin | | Authors: | Yu, J, Raia, P, Ghent, C.M, Raisch, T, Sadian, Y, Barford, D, Raunser, S, Morgan, D.O, Boland, A. | | Deposit date: | 2021-02-14 | | Release date: | 2021-08-04 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of human separase regulation by securin and CDK1-cyclin B1.
Nature, 596, 2021
|
|
5E78
 
 | | Crystal structure of P450 BM3 heme domain variant complexed with Co(III)Sep | | Descriptor: | 1,3,6,8,10,13,16,19-octaazabicyclo[6.6.6]icosane, Bifunctional P-450/NADPH-P450 reductase, CHLORIDE ION, ... | | Authors: | Panneerselvm, S, Shehzad, A, Bocola, M, Mueller-Dieckmann, J, Schwaneberg, U. | | Deposit date: | 2015-10-12 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic insights into a cobalt (III) sepulchrate based alternative cofactor system of P450 BM3 monooxygenase.
Biochim. Biophys. Acta, 1866, 2018
|
|
