6ZVS
 
 | | C12 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | Vipp1 | | Authors: | Liu, J, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
4HFX
 
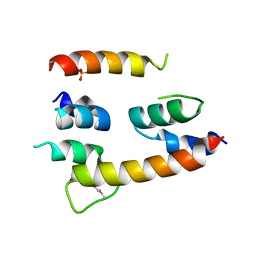 | | Crystal structure of a transcription elongation factor B polypeptide 3 from Homo sapiens, Northeast Structural Genomics consortium target id HR4748B. | | Descriptor: | SULFATE ION, Transcription elongation factor B polypeptide 3 | | Authors: | Seetharaman, J, Su, M, Ciccosanti, C, Sahdev, S, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-10-05 | | Release date: | 2012-12-12 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal structure of a transcription elongation factor B polypeptide 3 from Homo sapiens, Northeast Structural Genomics consortium target id HR4748B. (CASP Target)
TO BE PUBLISHED
|
|
6ZW6
 
 | | C16 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | vipp1 | | Authors: | Liu, J, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
4HG6
 
 | |
6ZW7
 
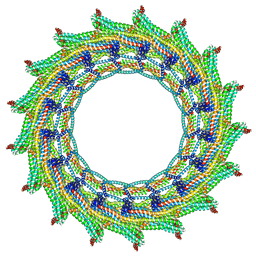 | | C17 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | vipp1 | | Authors: | Liu, J, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
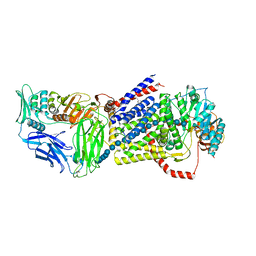
4GNG
 
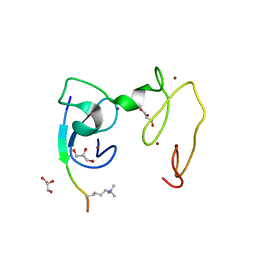 | | Crystal Structure of NSD3 tandem PHD5-C5HCH domains complexed with H3K9me3 peptide | | Descriptor: | GLYCEROL, Histone H3.3, Histone-lysine N-methyltransferase NSD3, ... | | Authors: | Li, F, He, C, Wu, J, Shi, Y. | | Deposit date: | 2012-08-17 | | Release date: | 2013-01-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The methyltransferase NSD3 has chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (nuclear receptor SET domain) family members in their histone H3 recognition.
J.Biol.Chem., 288, 2013
|
|
3PMI
 
 | | PWWP Domain of Human Mutated Melanoma-Associated Antigen 1 | | Descriptor: | DI(HYDROXYETHYL)ETHER, PWWP domain-containing protein MUM1, SULFATE ION, ... | | Authors: | Lam, R, Zeng, H, Loppnau, P, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-17 | | Release date: | 2010-12-15 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structural and histone binding ability characterizations of human PWWP domains.
Plos One, 6, 2011
|
|
6ZVV
 
 | | X-RAY STRUCTURE OF THE HALOALKANE DEHALOGENASE HALOTAG7-P174W LABELED WITH A CHLOROALKANE-TETRAMETHYLRHODAMINE FLUOROPHORE SUBSTRATE | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Tarnawski, M, Frei, M, Hiblot, J, Johnsson, K. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-RAY STRUCTURE OF THE HALOALKANE DEHALOGENASE HALOTAG7-P174W LABELED WITH A CHLOROALKANE-TETRAMETHYLRHODAMINE FLUOROPHORE SUBSTRATE
To Be Published
|
|
6ZVU
 
 | | X-RAY STRUCTURE OF THE HALOALKANE DEHALOGENASE HALOTAG7-P174L LABELED WITH A CHLOROALKANE-TETRAMETHYLRHODAMINE FLUOROPHORE SUBSTRATE | | Descriptor: | CHLORIDE ION, GLYCEROL, Haloalkane dehalogenase, ... | | Authors: | Tarnawski, M, Frei, M, Hiblot, J, Johnsson, K. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-RAY STRUCTURE OF THE HALOALKANE DEHALOGENASE HALOTAG7-P174L LABELED WITH A CHLOROALKANE-TETRAMETHYLRHODAMINE FLUOROPHORE SUBSTRATE
To Be Published
|
|
6ZVX
 
 | | X-RAY STRUCTURE OF THE HALOALKANE DEHALOGENASE HALOTAG7-Q165H-P174L LABELED WITH A CHLOROALKANE-TETRAMETHYLRHODAMINE FLUOROPHORE SUBSTRATE | | Descriptor: | CHLORIDE ION, GLYCEROL, Haloalkane dehalogenase, ... | | Authors: | Tarnawski, M, Frei, M, Hiblot, J, Johnsson, K. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-RAY STRUCTURE OF THE HALOALKANE DEHALOGENASE HALOTAG7-Q165H-P174L LABELED WITH A CHLOROALKANE-TETRAMETHYLRHODAMINE FLUOROPHORE SUBSTRATE
To Be Published
|
|
8G2J
 
 | | Hybrid aspen cellulose synthase-8 bound to UDP-glucose | | Descriptor: | Cellulose synthase, MAGNESIUM ION, URIDINE-5'-DIPHOSPHATE-GLUCOSE, ... | | Authors: | Verma, P, Zimmer, J. | | Deposit date: | 2023-02-04 | | Release date: | 2023-03-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Insights into substrate coordination and glycosyl transfer of poplar cellulose synthase-8.
Biorxiv, 2023
|
|
4C4J
 
 | | Structure-based design of orally bioavailable pyrrolopyridine inhibitors of the mitotic kinase MPS1 | | Descriptor: | DUAL SPECIFICITY PROTEIN KINASE TTK, tert-butyl 6-{[2-chloro-4-(1-methyl-1H-imidazol-5-yl)phenyl]amino}-2-(1-methyl-1H-pyrazol-4-yl)-1H-pyrrolo[3,2-c]pyridine-1-carboxylate | | Authors: | Naud, S, Westwood, I.M, Faisal, A, Sheldrake, P, Bavetsias, V, Atrash, B, Liu, M, Hayes, A, Schmitt, J, Wood, A, Choi, V, Boxall, K, Mak, G, Gurden, M, Valenti, M, de Haven Brandon, A, Henley, A, Baker, R, McAndrew, C, Matijssen, B, Burke, R, Eccles, S.A, Raynaud, F.I, Linardopoulos, S, van Montfort, R, Blagg, J. | | Deposit date: | 2013-09-05 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Design of Orally Bioavailable 1H-Pyrrolo[3, 2-C]Pyridine Inhibitors of the Mitotic Kinase Monopolar Spindle 1 (Mps1).
J.Med.Chem., 56, 2013
|
|
4NAX
 
 | | Crystal structure of glutathione transferase PPUT_1760 from Pseudomonas putida, target EFI-507288, with one glutathione disulfide bound per one protein subunit | | Descriptor: | FORMIC ACID, GLYCEROL, Glutathione S-transferase, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-22 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.301 Å) | | Cite: | Crystal structure of glutathione transferase Pput_1760 from Pseudomonas putida, target EFI-507288
To be Published
|
|
6ZVW
 
 | | X-RAY STRUCTURE OF THE HALOALKANE DEHALOGENASE HALOTAG7-Q165H LABELED WITH A CHLOROALKANE-TETRAMETHYLRHODAMINE FLUOROPHORE SUBSTRATE | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Tarnawski, M, Frei, M, Hiblot, J, Johnsson, K. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-RAY STRUCTURE OF THE HALOALKANE DEHALOGENASE HALOTAG7-Q165H LABELED WITH A CHLOROALKANE-TETRAMETHYLRHODAMINE FLUOROPHORE SUBSTRATE
To Be Published
|
|
2PHC
 
 | | Crystal structure of conserved uncharacterized protein PH0987 from Pyrococcus horikoshii | | Descriptor: | Uncharacterized protein PH0987 | | Authors: | Swindell II, J.T, Chen, L, Zhu, J, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Fu, Z.-Q, Chrzas, J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-10 | | Release date: | 2007-05-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of conserved uncharacterized protein PH0987 from Pyrococcus horikoshii.
To be Published
|
|
3BL8
 
 | | Crystal structure of the extracellular domain of neuroligin 2A from mouse | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Neuroligin-2, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jin, X, Koehnke, J, Shapiro, L. | | Deposit date: | 2007-12-10 | | Release date: | 2008-02-19 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the extracellular cholinesterase-like domain from neuroligin-2.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
8G27
 
 | | Hybrid aspen cellulose synthase-8 bound to UDP | | Descriptor: | Cellulose synthase, URIDINE-5'-DIPHOSPHATE, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Verma, P, Zimmer, J. | | Deposit date: | 2023-02-03 | | Release date: | 2023-03-01 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Insights into substrate coordination and glycosyl transfer of poplar cellulose synthase-8.
Biorxiv, 2023
|
|
2PH7
 
 | | Crystal structure of AF2093 from Archaeoglobus fulgidus | | Descriptor: | Uncharacterized protein AF_2093 | | Authors: | Chang, J.C, Yang, H, Hwang, J, Zhu, J, Chen, L, Fu, Z.-Q, Xu, H, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2007-04-10 | | Release date: | 2007-05-08 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of AF2093 from Archaeoglobus fulgidus.
To be Published
|
|
6ZZ2
 
 | | Cocktail experiment E: fragments 52, 58, and 63 at 90 mM concentration in complex with Endothiapepsin | | Descriptor: | DIMETHYL SULFOXIDE, Endothiapepsin, GLYCEROL, ... | | Authors: | Hassaan, E, Klebe, G, Heine, A, Schiebel, J, Koester, H. | | Deposit date: | 2020-08-03 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.14885437 Å) | | Cite: | Cocktail experiment E: fragments 52, 58, and 63 at 90 mM concentration in complex with Endothiapepsin
To Be Published
|
|
8C7B
 
 | |
6ZZ4
 
 | | Crystal structure of the PTPN2 C216G mutant | | Descriptor: | PHOSPHATE ION, Tyrosine-protein phosphatase non-receptor type 2 | | Authors: | Mechaly, A.E, Berthelet, J, Nian, Q, Parlato, M, Cerf-Bensussan, N, Haouz, A, Rodrigues-Lima, F. | | Deposit date: | 2020-08-03 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural characterization of a pathogenic mutant of human protein tyrosine phosphatase PTPN2 (Cys216Gly) that causes very early onset autoimmune enteropathy.
Protein Sci., 31, 2022
|
|
6ZYU
 
 | | Structure of the GluA2 ligand-binding domain (L483Y-N754S) in complex with glutamate and BPAM549 | | Descriptor: | (4~{R})-4-cyclopropyl-7-fluoranyl-3,4-dihydro-2~{H}-thiochromene 1,1-dioxide, (4~{S})-4-cyclopropyl-7-fluoranyl-3,4-dihydro-2~{H}-thiochromene 1,1-dioxide, ACETATE ION, ... | | Authors: | Dorosz, J, Christensen, K.M, Kastrup, J.S. | | Deposit date: | 2020-08-03 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Development of Thiochroman Dioxide Analogues of Benzothiadiazine Dioxides as New Positive Allosteric Modulators of alpha-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic Acid (AMPA) Receptors.
Acs Chem Neurosci, 12, 2021
|
|
4CBI
 
 | | Pestivirus NS3 helicase | | Descriptor: | SERINE PROTEASE NS3 | | Authors: | Tortorici, M.A, Duquerroy, S, Kwok, J, Vonrhein, C, Perez, J, Lamp, B, Bricogne, G, Rumenapf, T, Vachette, P, Rey, F.A. | | Deposit date: | 2013-10-14 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-Ray Structure of the Pestivirus Ns3 Helicase and its Conformation in Solution.
J.Virol., 89, 2015
|
|
8C7A
 
 | |
8BZQ
 
 | |
