1ZS3
 
 | | The crystal structure of the Lactococcus lactis MG1363 DpsB protein | | 分子名称: | Lactococcus lactis MG1363 DpsA | | 著者 | Stillman, T.J, Upadhyay, M, Norte, V.A, Sedelnikova, S.E, Carradus, M, Tzokov, S, Bullough, P.A, Shearman, C.A, Gasson, M.J, Williams, C.H, Artymiuk, P.J, Green, J. | | 登録日 | 2005-05-23 | | 公開日 | 2005-08-30 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | The crystal structures of Lactococcus lactis MG1363 Dps proteins reveal the presence of an N-terminal helix that is required for DNA binding.
Mol.Microbiol., 57, 2005
|
|
7L7L
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with NR01-129 | | 分子名称: | 1,1,1-trifluoro-2-methylpropan-2-yl [(2R,6S,12Z,13aS,14aR,16aS)-2-{[6-methoxy-3-(trifluoromethyl)quinoxalin-2-yl]oxy}-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, 1,2-ETHANEDIOL, NS3/4A protease, ... | | 著者 | Zephyr, J, Schiffer, C.A. | | 登録日 | 2020-12-29 | | 公開日 | 2021-09-01 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Discovery of Quinoxaline-Based P1-P3 Macrocyclic NS3/4A Protease Inhibitors with Potent Activity against Drug-Resistant Hepatitis C Virus Variants.
J.Med.Chem., 64, 2021
|
|
1R0E
 
 | | Glycogen synthase kinase-3 beta in complex with 3-indolyl-4-arylmaleimide inhibitor | | 分子名称: | 3-[3-(2,3-DIHYDROXY-PROPYLAMINO)-PHENYL]-4-(5-FLUORO-1-METHYL-1H-INDOL-3-YL)-PYRROLE-2,5-DIONE, CITRATE ANION, Glycogen synthase kinase-3 beta | | 著者 | Allard, J, Nikolcheva, T, Gong, L, Wang, J, Dunten, P, Avnur, Z, Waters, R, Sun, Q, Skinner, B. | | 登録日 | 2003-09-20 | | 公開日 | 2004-10-12 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | From genetics to therapeutics: the Wnt pathway and osteoporosis
To be Published
|
|
5CCB
 
 | | Crystal structure of human m1A58 methyltransferase in a complex with tRNA3Lys and SAH | | 分子名称: | S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, tRNA (adenine(58)-N(1))-methyltransferase catalytic subunit TRMT61A, ... | | 著者 | Finer-Moore, J, Czudnochowski, N, O'Connell III, J.D, Wang, A.L, Stroud, R.M. | | 登録日 | 2015-07-01 | | 公開日 | 2015-11-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure of the Human tRNA m(1)A58 Methyltransferase-tRNA3(Lys) Complex: Refolding of Substrate tRNA Allows Access to the Methylation Target.
J.Mol.Biol., 427, 2015
|
|
5COJ
 
 | | Structure of Hydroxyethylthiazole kinase ThiM from Staphylococcus aureus in complex with native substrate 2-(4-methyl-1,3-thiazol-5-yl)ethanol. | | 分子名称: | 2-(4-METHYL-THIAZOL-5-YL)-ETHANOL, Hydroxyethylthiazole kinase, MAGNESIUM ION | | 著者 | Drebes, J, Kuenz, M, Eberle, R.J, Oberthuer, D, Cang, H, Wrenger, C, Betzel, C. | | 登録日 | 2015-07-20 | | 公開日 | 2016-03-23 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of ThiM from Vitamin B1 biosynthetic pathway of Staphylococcus aureus - Insights into a novel pro-drug approach addressing MRSA infections.
Sci Rep, 6, 2016
|
|
7L8I
 
 | | SARS-CoV-2 Main Protease (Mpro) in Complex with Rupintrivir (P21) | | 分子名称: | 3C-like proteinase, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | 著者 | Lockbaum, G.J, Henes, M, Lee, J.M, Timm, J, Nalivaika, E.A, Yilmaz, N.K, Thompson, P.R, Schiffer, C.A. | | 登録日 | 2020-12-31 | | 公開日 | 2021-09-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Pan-3C Protease Inhibitor Rupintrivir Binds SARS-CoV-2 Main Protease in a Unique Binding Mode.
Biochemistry, 60, 2021
|
|
3T7I
 
 | | Crystal structure of Se-Met Rtt107p (residues 820-1070) | | 分子名称: | Regulator of Ty1 transposition protein 107 | | 著者 | Li, X, Li, F, Wu, J, Shi, Y. | | 登録日 | 2011-07-30 | | 公開日 | 2012-02-15 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of C-terminal Tandem BRCT Repeats of Rtt107 Protein Reveals Critical Role in Interaction with Phosphorylated Histone H2A during DNA Damage Repair
J.Biol.Chem., 287, 2012
|
|
5BYY
 
 | | ERK5 IN COMPLEX WITH SMALL MOLECULE | | 分子名称: | 2-{[2-ethoxy-4-(4-hydroxypiperidin-1-yl)phenyl]amino}-5,11-dimethyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Mitogen-activated protein kinase 7 | | 著者 | Chen, H, Tucker, J, Wang, X, Gavine, P.R, Philips, C, Augustin, M.A, Schreiner, P, Steinbacher, S, Preston, M, Ogg, D. | | 登録日 | 2015-06-11 | | 公開日 | 2016-05-04 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Discovery of a novel allosteric inhibitor-binding site in ERK5: comparison with the canonical kinase hinge ATP-binding site.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
7L8R
 
 | | Crystal structure of human GPX4-U46C mutant K48A | | 分子名称: | Isoform Cytoplasmic of Phospholipid hydroperoxide glutathione peroxidase, THIOCYANATE ION | | 著者 | Forouhar, F, Liu, H, Seibt, T, Saneto, R, Wigby, K, Friedman, J, Xia, X, Shchepinov, M.S, Ramesh, S, Conrad, M, Stockwell, B.R. | | 登録日 | 2020-12-31 | | 公開日 | 2021-12-29 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Patient-derived variant of GPX4 reveals the structural basis for its catalytic activity and degradation mechanism
Nat.Chem.Biol., 2021
|
|
5CIS
 
 | | The CUB1-EGF-CUB2 domains of rat MBL-associated serine protease-2 (MASP-2) bound to Ca2+ | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Mannan-binding lectin serine peptidase 2 | | 著者 | Nan, R, Furze, C.M, Wright, D.W, Gor, J, Wallis, R, Perkins, S.J. | | 登録日 | 2015-07-13 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | Flexibility in Mannan-Binding Lectin-Associated Serine Proteases-1 and -2 Provides Insight on Lectin Pathway Activation.
Structure, 25, 2017
|
|
7L8L
 
 | | Crystal structure of human R152H GPX4-U46C | | 分子名称: | Phospholipid hydroperoxide glutathione peroxidase, THIOCYANATE ION | | 著者 | Forouhar, F, Liu, H, Seibt, T, Saneto, R, Wigby, K, Friedman, J, Xia, X, Shchepinov, M.S, Ramesh, S, Conrad, M, Stockwell, B.R. | | 登録日 | 2020-12-31 | | 公開日 | 2021-12-29 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | Patient-derived variant of GPX4 reveals the structural basis for its catalytic activity and degradation mechanism
Nat.Chem.Biol., 2021
|
|
7L8M
 
 | | Crystal structure of human GPX4-U46C mutant K48L | | 分子名称: | Phospholipid hydroperoxide glutathione peroxidase | | 著者 | Forouhar, F, Liu, H, Seibt, T, Saneto, R, Friedman, J, Xia, X, Shchepinov, M.S, Ramesh, S, Conrad, M, Stockwell, B.R. | | 登録日 | 2020-12-31 | | 公開日 | 2021-12-29 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Patient-derived variant of GPX4 reveals the structural basis for its catalytic activity and degradation mechanism
Nat.Chem.Biol., 2021
|
|
4QB4
 
 | |
5C3O
 
 | | Crystal structure of the C-terminal truncated Neurospora crassa T7H (NcT7HdeltaC) in apo form | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, Thymine dioxygenase | | 著者 | Li, W, Zhang, T, Ding, J. | | 登録日 | 2015-06-17 | | 公開日 | 2015-10-21 | | 最終更新日 | 2015-12-02 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Molecular basis for the substrate specificity and catalytic mechanism of thymine-7-hydroxylase in fungi
Nucleic Acids Res., 43, 2015
|
|
5C3R
 
 | | Crystal structure of the full-length Neurospora crassa T7H in complex with alpha-KG and 5-hydroxymethyluracil (5hmU) | | 分子名称: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, 5-HYDROXYMETHYL URACIL, ... | | 著者 | Li, W, Zhang, T, Ding, J. | | 登録日 | 2015-06-17 | | 公開日 | 2015-10-21 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Molecular basis for the substrate specificity and catalytic mechanism of thymine-7-hydroxylase in fungi
Nucleic Acids Res., 43, 2015
|
|
5CKM
 
 | | The CUB1-EGF-CUB2 domains of rat MBL-associated serine protease-2 (MASP-2) bound to Ca2+ | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Mannan-binding lectin serine peptidase 2 | | 著者 | Nan, R, Furze, C.M, Wright, D.W, Gor, J, Wallis, R, Perkins, S.J. | | 登録日 | 2015-07-15 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.73 Å) | | 主引用文献 | Flexibility in Mannan-Binding Lectin-Associated Serine Proteases-1 and -2 Provides Insight on Lectin Pathway Activation.
Structure, 25, 2017
|
|
7LA4
 
 | | Integrin AlphaIIbBeta3-PT25-2 Complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Bush, M.W, Walz, T, Coller, B, Filizola, M, Spasic, A, Nesic, D, Li, J. | | 登録日 | 2021-01-05 | | 公開日 | 2022-01-12 | | 最終更新日 | 2022-07-27 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Electron microscopy shows that binding of monoclonal antibody PT25-2 primes integrin alpha IIb beta 3 for ligand binding.
Blood Adv, 5, 2021
|
|
5CM5
 
 | | Structure of Hydroxyethylthiazole Kinase ThiM from Staphylococcus aureus | | 分子名称: | Hydroxyethylthiazole kinase | | 著者 | Drebes, J, Kuenz, M, Eberle, R.J, Oberthuer, D, Cang, H, Wrenger, C, Betzel, C. | | 登録日 | 2015-07-16 | | 公開日 | 2016-03-23 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Structure of ThiM from Vitamin B1 biosynthetic pathway of Staphylococcus aureus - Insights into a novel pro-drug approach addressing MRSA infections.
Sci Rep, 6, 2016
|
|
4QN4
 
 | | Crystal structure of Neuraminidase N6 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, GLYCEROL, ... | | 著者 | Sun, X, Li, Q, Wu, Y, Liu, Y, Qi, J, Vavricka, C.J, Gao, G.F. | | 登録日 | 2014-06-17 | | 公開日 | 2014-07-30 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of influenza virus N7: the last piece of the neuraminidase "jigsaw" puzzle.
J.Virol., 88, 2014
|
|
5C74
 
 | |
4D90
 
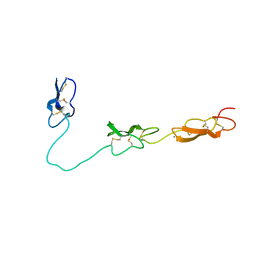 | | Crystal Structure of Del-1 EGF domains | | 分子名称: | 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Chen, Q, Schurpf, T, Springer, T, Wang, J. | | 登録日 | 2012-01-11 | | 公開日 | 2012-05-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.601 Å) | | 主引用文献 | The RGD finger of Del-1 is a unique structural feature critical for integrin binding.
Faseb J., 26, 2012
|
|
1QN1
 
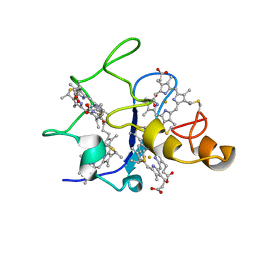 | | SOLUTION STRUCTURE OF DESULFOVIBRIO GIGAS FERRICYTOCHROME C3, NMR, 15 STRUCTURES | | 分子名称: | CYTOCHROME C3, HEME C | | 著者 | Brennan, L, Messias, A.C, Legall, J, Turner, D.L, Xavier, A.V. | | 登録日 | 1999-10-11 | | 公開日 | 2000-10-12 | | 最終更新日 | 2019-11-06 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Basis for the Network of Functional Cooperativities in Cytochromes C3 from Desulfovibrio Gigas: Solution Structures of the Oxidised and Reduced States
J.Mol.Biol., 298, 2000
|
|
1UN2
 
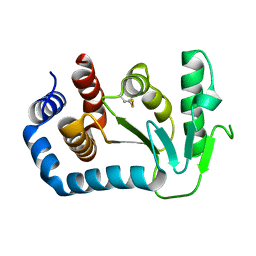 | | Crystal structure of circularly permuted CPDSBA_Q100T99: Preserved Global Fold and Local Structural Adjustments | | 分子名称: | THIOL-DISULFIDE INTERCHANGE PROTEIN | | 著者 | Manjasetty, B.A, Hennecke, J, Glockshuber, R, Heinemann, U. | | 登録日 | 2003-09-03 | | 公開日 | 2003-09-26 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure of Circularly Permuted Dsba(Q100T99): Preserved Global Fold and Local Structural Adjustments
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1UT1
 
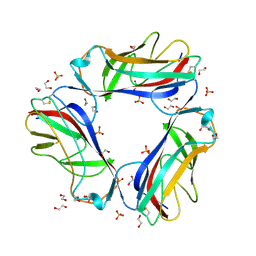 | | DraE adhesin from Escherichia Coli | | 分子名称: | 1,2-ETHANEDIOL, DR HEMAGGLUTININ STRUCTURAL SUBUNIT, SULFATE ION | | 著者 | Anderson, K.L, Billington, J, Pettigrew, D, Cota, E, Roversi, P, Simpson, P, Chen, H.A, Urvil, P, Dumerle, L, Barlow, P, Medof, E, Smith, R.A.G, Nowicki, B, Le Bouguenec, C, Lea, S.M, Matthews, S. | | 登録日 | 2003-12-02 | | 公開日 | 2004-08-31 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | High Resolution Studies of the Afa/Dr Adhesin Drae and its Interaction with Chloramphenicol
J.Biol.Chem., 279, 2004
|
|
5C9F
 
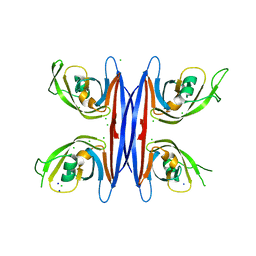 | | Crystal structure of a retropepsin-like aspartic protease from Rickettsia conorii | | 分子名称: | ApRick protease, CHLORIDE ION, SODIUM ION | | 著者 | Li, M, Gustchina, A, Cruz, R, Simoes, M, Curto, P, Martinez, J, Faro, C, Simoes, I, Wlodawer, A. | | 登録日 | 2015-06-26 | | 公開日 | 2015-10-14 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of RC1339/APRc from Rickettsia conorii, a retropepsin-like aspartic protease.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
