8FSK
 
 | | LSD1-CoREST in complex with T18, short soaking | | 分子名称: | Lysine-specific histone demethylase 1A, REST corepressor 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl (2R,3S,4S)-5-[(1R,3S,3aS,13R)-3-(3-benzamidophenyl)-1-hydroxy-10,11-dimethyl-4,6-dioxo-2,3,5,6-tetrahydro-1H-benzo[g]pyrrolo[2,1-e]pteridin-8(4H)-yl]-2,3,4-trihydroxypentyl dihydrogen diphosphate (non-preferred name) | | 著者 | Caroli, J, Mattevi, A. | | 登録日 | 2023-01-10 | | 公開日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (3.13 Å) | | 主引用文献 | Distal drug resistance mutations promote covalent inhibitor-adduct Grob fragmentation in LSD1
To Be Published
|
|
7AD1
 
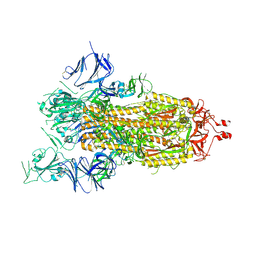 | | Cryo-EM structure of a prefusion stabilized SARS-CoV-2 Spike (D614N, R682S, R685G, A892P, A942P and V987P)(One up trimer) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Envelope glycoprotein,Spike glycoprotein,Envelope glycoprotein,SARS-CoV-2 S protein | | 著者 | Rutten, L, Renault, L.L.R, Juraszek, J, Langedijk, J.P.M. | | 登録日 | 2020-09-14 | | 公開日 | 2020-11-04 | | 最終更新日 | 2021-01-27 | | 実験手法 | ELECTRON MICROSCOPY (2.92 Å) | | 主引用文献 | Stabilizing the closed SARS-CoV-2 spike trimer.
Nat Commun, 12, 2021
|
|
5Y88
 
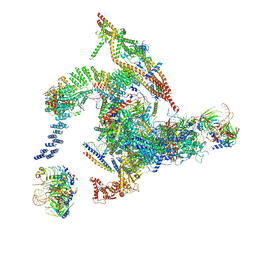 | | Cryo-EM structure of the intron-lariat spliceosome ready for disassembly from S.cerevisiae at 3.5 angstrom | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, Intron lariat, ... | | 著者 | Wan, R, Yan, C, Bai, R, Lei, J, Shi, Y. | | 登録日 | 2017-08-20 | | 公開日 | 2018-08-01 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.46 Å) | | 主引用文献 | Structure of an Intron Lariat Spliceosome from Saccharomyces cerevisiae
Cell(Cambridge,Mass.), 171, 2017
|
|
8FZH
 
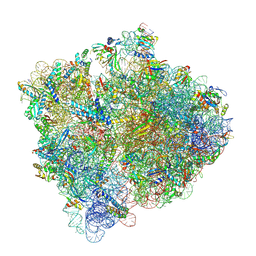 | | Cryo-EM structure of an E. coli non-rotated ribosome termination complex bound with RF1, P- and E-site tRNAPhe (State II-D) | | 分子名称: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Rybak, M.Y, Li, L, Lin, J, Gagnon, M.G. | | 登録日 | 2023-01-28 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-07-31 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | The ribosome termination complex remodels release factor RF3 and ejects GDP.
Nat.Struct.Mol.Biol., 2024
|
|
5YDX
 
 | |
1H5Q
 
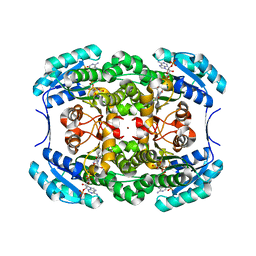 | | Mannitol dehydrogenase from Agaricus bisporus | | 分子名称: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-DEPENDENT MANNITOL DEHYDROGENASE, NICKEL (II) ION | | 著者 | Horer, S, Stoop, J, Mooibroek, H, Baumann, U, Sassoon, J. | | 登録日 | 2001-05-24 | | 公開日 | 2001-06-21 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The Crystallographic Structure of the Mannitol 2-Dehydrogenase Nadp+ Binary Complex from Agaricus Bisporus
J.Biol.Chem., 276, 2001
|
|
6ZQV
 
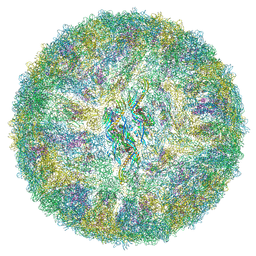 | | Cryo-EM structure of mature Spondweni virus | | 分子名称: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Genome polyprotein | | 著者 | Renner, M, Dejnirattisai, W, Carrique, L, Serna Martin, I, Karia, D, Ilca, S.L, Ho, S.F, Kotecha, A, Keown, J.R, Mongkolsapaya, J, Screaton, G.R, Grimes, J.M. | | 登録日 | 2020-07-10 | | 公開日 | 2021-01-20 | | 最終更新日 | 2021-08-04 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Flavivirus maturation leads to the formation of an occupied lipid pocket in the surface glycoproteins.
Nat Commun, 12, 2021
|
|
8CJ1
 
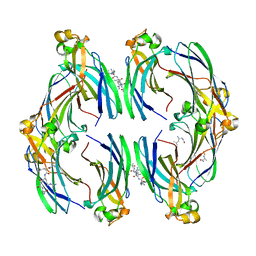 | | Urea-based foldamer inhibitor c3u_3 chimera in complex with ASF1 histone chaperone | | 分子名称: | Histone chaperone ASF1A, c3u_3 chimera inhibitor of histone chaperone ASF1 | | 著者 | Perrin, M.E, Li, B, Mbianda, J, Ropars, V, Legrand, P, Douat, C, Ochsenbein, F, Guichard, G. | | 登録日 | 2023-02-11 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.564 Å) | | 主引用文献 | Unexpected binding modes of inhibitors to the histone chaperone ASF1 revealed by a foldamer scanning approach.
Chem.Commun.(Camb.), 59, 2023
|
|
6ZQG
 
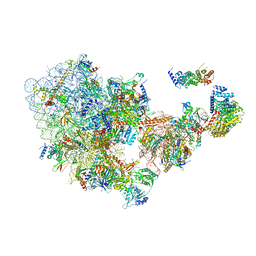 | | Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae, state Dis-C | | 分子名称: | 18S rRNA, 40S ribosomal protein S1-A, 40S ribosomal protein S11-A, ... | | 著者 | Cheng, J, Lau, B, Venuta, G.L, Berninghausen, O, Hurt, E, Beckmann, R. | | 登録日 | 2020-07-09 | | 公開日 | 2020-09-23 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | 90 S pre-ribosome transformation into the primordial 40 S subunit.
Science, 369, 2020
|
|
8HGU
 
 | |
8CJ2
 
 | | Urea-based foldamer inhibitor c3u_5 chimera in complex with ASF1 histone chaperone | | 分子名称: | GLYCEROL, Histone chaperone ASF1A, SULFATE ION, ... | | 著者 | Perrin, M.E, Li, B, Mbianda, J, Ropars, V, Legrand, P, Douat, C, Ochsenbein, F, Guichard, G. | | 登録日 | 2023-02-11 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (2.127 Å) | | 主引用文献 | Unexpected binding modes of inhibitors to the histone chaperone ASF1 revealed by a foldamer scanning approach.
Chem.Commun.(Camb.), 59, 2023
|
|
3O5C
 
 | | Cytochrome c Peroxidase BccP of Shewanella oneidensis | | 分子名称: | (R,R)-2,3-BUTANEDIOL, CALCIUM ION, Cytochrome c551 peroxidase, ... | | 著者 | Seidel, J, Einsle, O. | | 登録日 | 2010-07-28 | | 公開日 | 2011-08-03 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Investigation of the Electron Transport Chain to and the Catalytic Activity of the Diheme Cytochrome c Peroxidase CcpA of Shewanella oneidensis.
Appl.Environ.Microbiol., 77, 2011
|
|
6ZXH
 
 | | Cryo-EM structure of a late human pre-40S ribosomal subunit - State H2 | | 分子名称: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | 著者 | Ameismeier, M, Zemp, I, van den Heuvel, J, Thoms, M, Berninghausen, O, Kutay, U, Beckmann, R. | | 登録日 | 2020-07-29 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Structural basis for the final steps of human 40S ribosome maturation.
Nature, 587, 2020
|
|
5YK6
 
 | | Crystal Structure of Mmm1 | | 分子名称: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, Maintenance of mitochondrial morphology protein 1 | | 著者 | Jeong, H, Park, J, Lee, C. | | 登録日 | 2017-10-12 | | 公開日 | 2018-01-10 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structures of Mmm1 and Mdm12-Mmm1 reveal mechanistic insight into phospholipid trafficking at ER-mitochondria contact sites.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5YDH
 
 | | Crystal structure of acetylcholinesterase catalytic subunits of the malaria vector anopheles gambiae, 3.2 A | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Acetylcholinesterase, ... | | 著者 | Han, Q, Guan, H, Robinson, H, Ding, H, Liao, C, Li, J. | | 登録日 | 2017-09-13 | | 公開日 | 2018-03-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.21 Å) | | 主引用文献 | Crystal structures of acetylcholinesterase of the malaria vector Anopheles gambiae reveal a polymerization interface, ligand binding residues and post translational modifications
To Be Published
|
|
5YKI
 
 | | Crystal structure of the engineered nine-repeat PUF domain in complex with cognate 9nt-RNA | | 分子名称: | Pumilio homolog 1, RNA (5'-R(*UP*GP*UP*UP*GP*UP*AP*UP*A)-3') | | 著者 | Zhao, Y.Y, Wang, J, Li, H.T, Wang, Z.X, Wu, J.W. | | 登録日 | 2017-10-14 | | 公開日 | 2018-03-14 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Expanding RNA binding specificity and affinity of engineered PUF domains.
Nucleic Acids Res., 46, 2018
|
|
5YKN
 
 | | crystal structure of Arabidopsis thaliana JMJ14 catalytic domain | | 分子名称: | NICKEL (II) ION, Probable lysine-specific demethylase JMJ14, ZINC ION | | 著者 | Yang, Z, Du, J. | | 登録日 | 2017-10-15 | | 公開日 | 2017-12-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of the Arabidopsis JMJ14-H3K4me3 Complex Provides Insight into the Substrate Specificity of KDM5 Subfamily Histone Demethylases.
Plant Cell, 30, 2018
|
|
6ZMT
 
 | | SARS-CoV-2 Nsp1 bound to a pre-40S-like ribosome complex | | 分子名称: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | 著者 | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | 登録日 | 2020-07-03 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
5YEH
 
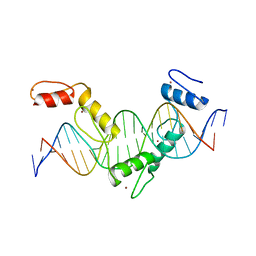 | | Crystal structure of CTCF ZFs4-8-eCBS | | 分子名称: | DNA (5'-D(*AP*CP*GP*GP*TP*TP*TP*CP*CP*GP*CP*TP*AP*GP*AP*GP*GP*GP*CP*G)-3'), DNA (5'-D(*TP*CP*GP*CP*CP*CP*TP*CP*TP*AP*GP*CP*GP*GP*AP*AP*AP*CP*CP*G)-3'), Transcriptional repressor CTCF, ... | | 著者 | Yin, M, Wang, J, Wang, M, Li, X, Wang, Y. | | 登録日 | 2017-09-17 | | 公開日 | 2017-11-29 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.328 Å) | | 主引用文献 | Molecular mechanism of directional CTCF recognition of a diverse range of genomic sites
Cell Res., 27, 2017
|
|
8HM5
 
 | |
5YER
 
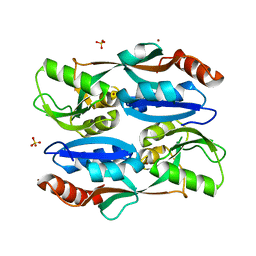 | | Regulatory domain of HypT from Salmonella typhimurium (Bromide ion-bound) | | 分子名称: | BROMIDE ION, Cell density-dependent motility repressor, SULFATE ION | | 著者 | Jo, I, Hong, S, Ahn, J, Ha, N.C. | | 登録日 | 2017-09-19 | | 公開日 | 2018-11-28 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.301 Å) | | 主引用文献 | Structural basis for HOCl recognition and regulation mechanisms of HypT, a hypochlorite-specific transcriptional regulator.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
8AKK
 
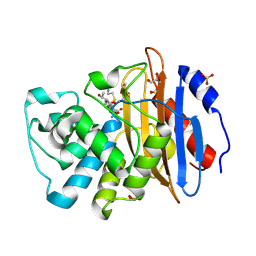 | | Acyl-enzyme complex of imipenem bound to deacylation mutant KPC-2 (E166Q) | | 分子名称: | (2R,4S)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-[(2-{[(Z)-iminomethyl]amino}ethyl)sulfanyl]-3,4-dihydro-2H-pyrrole-5-ca rboxylic acid, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL, ... | | 著者 | Tooke, C.L, Hinchliffe, P, Spencer, J. | | 登録日 | 2022-07-29 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | Tautomer-Specific Deacylation and Omega-Loop Flexibility Explain the Carbapenem-Hydrolyzing Broad-Spectrum Activity of the KPC-2 beta-Lactamase.
J.Am.Chem.Soc., 145, 2023
|
|
8AKI
 
 | | Acyl-enzyme complex of ampicillin bound to deacylation mutant KPC-2 (E166Q) | | 分子名称: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL, ... | | 著者 | Tooke, C.L, Hinchliffe, P, Spencer, J. | | 登録日 | 2022-07-29 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Tautomer-Specific Deacylation and Omega-Loop Flexibility Explain the Carbapenem-Hydrolyzing Broad-Spectrum Activity of the KPC-2 beta-Lactamase.
J.Am.Chem.Soc., 145, 2023
|
|
7A9D
 
 | | Crystal structure of H12 Haemagglutinin | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | 著者 | Xiong, X, Walker, P, Zhang, J, Gamblin, S, Skehel, J.J. | | 登録日 | 2020-09-01 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Hemagglutinin Structure and Activities.
Cold Spring Harb Perspect Med, 11, 2021
|
|
8AKL
 
 | | Acyl-enzyme complex of meropenem bound to deacylation mutant KPC-2 (E166Q) | | 分子名称: | (2S,3R,4R)-4-[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl-3-methyl-2-[(2S,3R)-3-oxidanyl-1-oxidanylidene-butan-2-yl]-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL, ... | | 著者 | Tooke, C.L, Hinchliffe, P, Spencer, J. | | 登録日 | 2022-07-29 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Tautomer-Specific Deacylation and Omega-Loop Flexibility Explain the Carbapenem-Hydrolyzing Broad-Spectrum Activity of the KPC-2 beta-Lactamase.
J.Am.Chem.Soc., 145, 2023
|
|
