8GZY
 
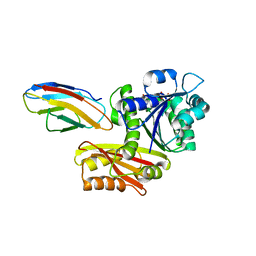 | | Escherichia coli FtsZ complexed with monobody (P21) | | 分子名称: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, Monobody | | 著者 | Matsumura, H, Yoshizawa, T, Fujita, J, Tanaka, S, Amesaka, H. | | 登録日 | 2022-09-27 | | 公開日 | 2023-07-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
5YEZ
 
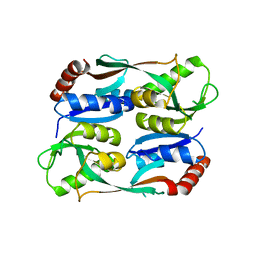 | | Regulatory domain of HypT M206Q mutant from Salmonella typhimurium | | 分子名称: | Cell density-dependent motility repressor | | 著者 | Jo, I, Hong, S, Ahn, J, Ha, N.C. | | 登録日 | 2017-09-20 | | 公開日 | 2018-10-03 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis for HOCl recognition and regulation mechanisms of HypT, a hypochlorite-specific transcriptional regulator.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
2PH5
 
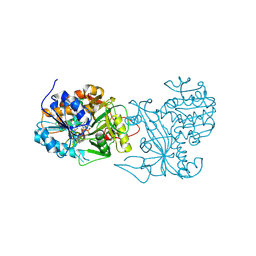 | | Crystal structure of the homospermidine synthase hss from Legionella pneumophila in complex with NAD, Northeast Structural Genomics Target LgR54 | | 分子名称: | Homospermidine synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | 著者 | Forouhar, F, Hussain, M, Seetharaman, J, Fang, Y, Janjua, H, Xiao, R, Cunningham, K, Ma, L.-C, Owens, L, Liu, J, Baran, M.C, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2007-04-10 | | 公開日 | 2007-05-15 | | 最終更新日 | 2018-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of the homospermidine synthase hss from Legionella pneumophila in complex with NAD.
To be Published
|
|
8H0I
 
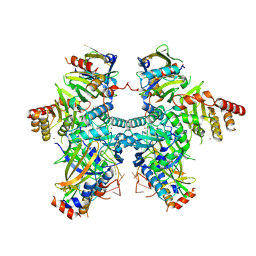 | | Cryo-EM structure of APOBEC3G-Vif complex | | 分子名称: | APOBEC3G, CHLORIDE ION, Core binding factor beta, ... | | 著者 | Kouno, T, Shibata, S, Hyun, J, Kim, T.G, Wolf, M. | | 登録日 | 2022-09-29 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structural insights into RNA bridging between HIV-1 Vif and antiviral factor APOBEC3G.
Nat Commun, 14, 2023
|
|
6ZQA
 
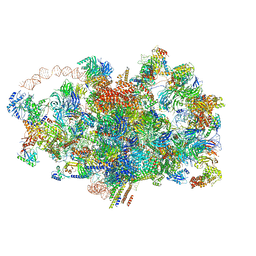 | | Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae, state A (Poly-Ala) | | 分子名称: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | 著者 | Cheng, J, Lau, B, Venuta, G.L, Berninghausen, O, Hurt, E, Beckmann, R. | | 登録日 | 2020-07-09 | | 公開日 | 2020-09-23 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | 90 S pre-ribosome transformation into the primordial 40 S subunit.
Science, 369, 2020
|
|
7D90
 
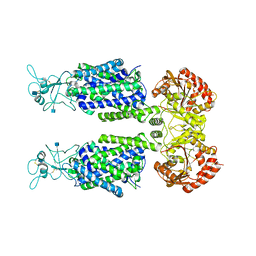 | | human potassium-chloride co-transporter KCC3 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, potassium-chloride cotransporter 3 | | 著者 | Xie, Y, Chang, S, Zhao, C, Ye, S, Guo, J. | | 登録日 | 2020-10-12 | | 公開日 | 2020-12-30 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structures and an activation mechanism of human potassium-chloride cotransporters.
Sci Adv, 6, 2020
|
|
7D8Z
 
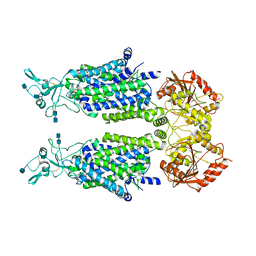 | | human potassium-chloride co-transporter KCC2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, potassium-chloride cotransporter 2 | | 著者 | Xie, Y, Chang, S, Zhao, C, Ye, S, Guo, J. | | 登録日 | 2020-10-11 | | 公開日 | 2020-12-30 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structures and an activation mechanism of human potassium-chloride cotransporters.
Sci Adv, 6, 2020
|
|
8HH2
 
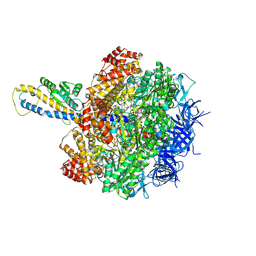 | | F1 domain of FoF1-ATPase from Bacillus PS3,post-hyd,highATP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | 著者 | Nakano, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | 登録日 | 2022-11-16 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Rotation mechanism of ATP synthases driven by ATP hydrolysis
To Be Published
|
|
3J0P
 
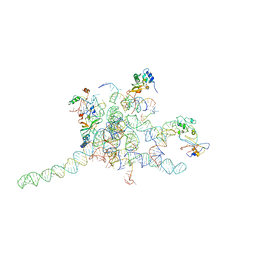 | | Core of mammalian 80S pre-ribosome in complex with tRNAs fitted to a 10.6A cryo-em map: rotated PRE state 1 | | 分子名称: | 40S ribosomal RNA fragment, 60S ribosomal RNA fragment, Ribosomal protein L10a, ... | | 著者 | Budkevich, T, Giesebrecht, J, Altman, R, Munro, J, Mielke, T, Nierhaus, K, Blanchard, S, Spahn, C.M. | | 登録日 | 2011-10-06 | | 公開日 | 2011-11-16 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (10.6 Å) | | 主引用文献 | Structure and dynamics of the Mammalian ribosomal pretranslocation complex.
Mol.Cell, 44, 2011
|
|
4J8B
 
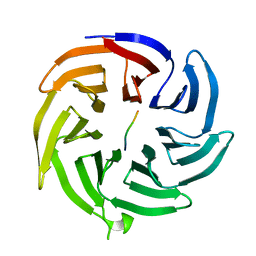 | |
6ZTQ
 
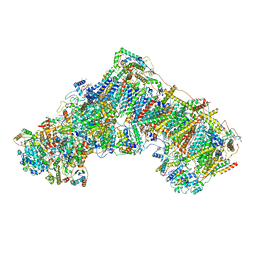 | | Cryo-EM structure of respiratory complex I from Mus musculus inhibited by piericidin A at 3.0 A | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Bridges, H.R, Blaza, J.N, Agip, A.N.A, Hirst, J. | | 登録日 | 2020-07-20 | | 公開日 | 2020-10-21 | | 最終更新日 | 2020-10-28 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structure of inhibitor-bound mammalian complex I.
Nat Commun, 11, 2020
|
|
2PIH
 
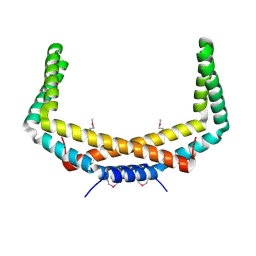 | | Crystal structure of Protein ymcA from Bacillus subtilis, NorthEast Structural Genomics target SR375 | | 分子名称: | Protein ymcA | | 著者 | Seetharaman, J, Abashidze, M, Forouhar, F, Wang, D, Fang, Y, Cunningham, K, Ma, L.-C, Xia, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2007-04-13 | | 公開日 | 2007-05-01 | | 最終更新日 | 2018-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of Protein ymcA from Bacillus subtilis.
To be Published
|
|
8HHC
 
 | | F1 domain of FoF1-ATPase from Bacillus PS3,post-hyd',lowATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ATP synthase subunit alpha, ... | | 著者 | Nakano, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | 登録日 | 2022-11-16 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Rotation mechanism of ATP synthases driven by ATP hydrolysis
To Be Published
|
|
8HH4
 
 | | F1 domain of FoF1-ATPase from Bacillus PS3,101 degrees, highATP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | 著者 | Nakano, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | 登録日 | 2022-11-16 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Rotation mechanism of ATP synthases driven by ATP hydrolysis
To Be Published
|
|
8HHB
 
 | | F1 domain of FoF1-ATPase from Bacillus PS3,step waiting,lowATP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | 著者 | Nakano, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | 登録日 | 2022-11-16 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Rotation mechanism of ATP synthases driven by ATP hydrolysis
To Be Published
|
|
6ZXT
 
 | | High resolution crystal structure of chloroplastic ribose-5-phosphate isomerase from Chlamydomonas reinhardtii | | 分子名称: | Ribose-5-phosphate isomerase, SODIUM ION, SULFATE ION | | 著者 | Le Moigne, T, Crozet, P, Lemaire, S.D, Henri, J. | | 登録日 | 2020-07-30 | | 公開日 | 2020-11-04 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | High-Resolution Crystal Structure of Chloroplastic Ribose-5-Phosphate Isomerase from Chlamydomonas reinhardtii -An Enzyme Involved in the Photosynthetic Calvin-Benson Cycle.
Int J Mol Sci, 21, 2020
|
|
8HH8
 
 | | F1 domain of FoF1-ATPase from Bacillus PS3,post-hyd,lowATP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | 著者 | Nakano, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | 登録日 | 2022-11-16 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Rotation mechanism of ATP synthases driven by ATP hydrolysis
To Be Published
|
|
5YDW
 
 | | Full-length structure of HypT from Salmonella typhimuriuma (hypochlorite-specific LysR-type transcriptional regulator) | | 分子名称: | Cell density-dependent motility repressor | | 著者 | Jo, I, Hong, S, Ahn, J, Ha, N.C. | | 登録日 | 2017-09-15 | | 公開日 | 2018-11-28 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structural basis for HOCl recognition and regulation mechanisms of HypT, a hypochlorite-specific transcriptional regulator.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
8HHA
 
 | | F1 domain of FoF1-ATPase from Bacillus PS3,120 degrees,lowATP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | 著者 | Nakano, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | 登録日 | 2022-11-16 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Rotation mechanism of ATP synthases driven by ATP hydrolysis
To Be Published
|
|
5YEG
 
 | | Crystal structure of CTCF ZFs4-8-Hs5-1a complex | | 分子名称: | DNA (5'-D(*AP*CP*TP*TP*TP*AP*AP*CP*CP*AP*GP*CP*AP*GP*AP*GP*GP*GP*CP*G)-3'), DNA (5'-D(*TP*CP*GP*CP*CP*CP*TP*CP*TP*GP*CP*TP*GP*GP*TP*TP*AP*AP*AP*G)-3'), Transcriptional repressor CTCF, ... | | 著者 | Yin, M, Wang, J, Wang, M, Li, X. | | 登録日 | 2017-09-17 | | 公開日 | 2017-11-29 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Molecular mechanism of directional CTCF recognition of a diverse range of genomic sites
Cell Res., 27, 2017
|
|
8HH1
 
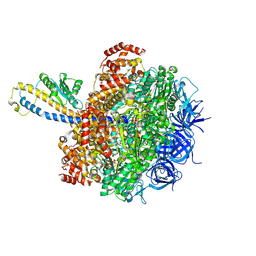 | | FoF1-ATPase from Bacillus PS3, 81 degrees, highATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ATP synthase subunit alpha, ... | | 著者 | Nakano, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | 登録日 | 2022-11-16 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Rotation mechanism of ATP synthases driven by ATP hydrolysis
To Be Published
|
|
6ZYO
 
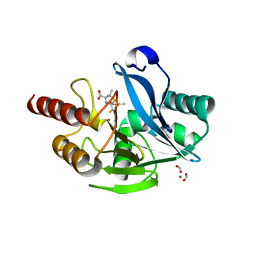 | | Structure of VIM-2 with 2-Mercaptomethyl-thiazolidine D-syn-1b | | 分子名称: | (2~{S},4~{S})-2-ethoxycarbonyl-5,5-dimethyl-2-(sulfanylmethyl)-1,3-thiazolidine-4-carboxylic acid, Beta-lactamase VIM-2, FORMIC ACID, ... | | 著者 | Hinchliffe, P, Spencer, J. | | 登録日 | 2020-08-02 | | 公開日 | 2021-01-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | 2-Mercaptomethyl-thiazolidines use conserved aromatic-S interactions to achieve broad-range inhibition of metallo-beta-lactamases.
Chem Sci, 12, 2021
|
|
8H1M
 
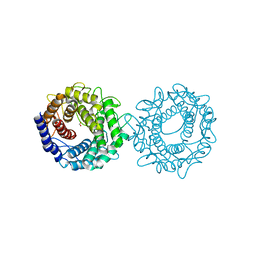 | | Crystal structure of glucose-2-epimerase mutant_D254A from Runella slithyformis Runsl_4512 | | 分子名称: | FORMIC ACID, N-acylglucosamine 2-epimerase | | 著者 | Wang, H, Sun, X.M, Saburi, W, Yu, J, Yao, M. | | 登録日 | 2022-10-03 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural insights into the substrate specificity and activity of a novel mannose 2-epimerase from Runella slithyformis.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
4JA8
 
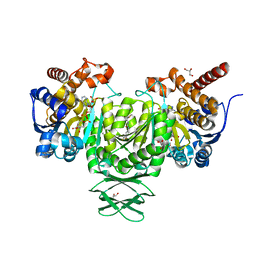 | | Complex of Mitochondrial Isocitrate Dehydrogenase R140Q Mutant with AGI-6780 Inhibitor | | 分子名称: | 1-[5-(cyclopropylsulfamoyl)-2-thiophen-3-yl-phenyl]-3-[3-(trifluoromethyl)phenyl]urea, CALCIUM ION, GLYCEROL, ... | | 著者 | Wei, W, Chen, L, Wu, M, Jiang, F, Travins, J, Qian, K, DeLaBarre, B. | | 登録日 | 2013-02-18 | | 公開日 | 2013-04-10 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation.
Science, 340, 2013
|
|
8H1N
 
 | | Crystal structure of glucose-2-epimerase mutant_D254A in complex with D-Glucitol from Runella slithyformis Runsl_4512 | | 分子名称: | FORMIC ACID, N-acylglucosamine 2-epimerase, sorbitol | | 著者 | Wang, H, Sun, X.M, Saburi, W, Yu, J, Yao, M. | | 登録日 | 2022-10-03 | | 公開日 | 2023-07-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Structural insights into the substrate specificity and activity of a novel mannose 2-epimerase from Runella slithyformis.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
