1B2I
 
 | |
3C7Q
 
 | | Structure of VEGFR2 kinase domain in complex with BIBF1120 | | Descriptor: | SULFATE ION, Vascular endothelial growth factor receptor 2, methyl (3Z)-3-{[(4-{methyl[(4-methylpiperazin-1-yl)acetyl]amino}phenyl)amino](phenyl)methylidene}-2-oxo-2,3-dihydro-1H-indole-6-carboxylate | | Authors: | Hilberg, F, Roth, G.J, Krssak, M, Kautschitsch, S, Sommergruber, W, Tontsch-Grunt, U, Garin-Chesa, P, Bader, G, Zoephel, A, Quant, J, Heckel, A, Rettig, W.J. | | Deposit date: | 2008-02-08 | | Release date: | 2008-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | BIBF 1120: triple angiokinase inhibitor with sustained receptor blockade and good antitumor efficacy.
Cancer Res., 68, 2008
|
|
1MX8
 
 | |
3C85
 
 | |
2VWM
 
 | | Aminopyrrolidine Factor Xa inhibitor | | Descriptor: | (4R)-4-{[(5-chlorothiophen-2-yl)carbonyl]amino}-N-(cyclopropylmethyl)-1-(2-{[2-fluoro-4-(2-oxopyridin-1(2H)-yl)phenyl]amino}-2-oxoethyl)-L-prolinamide, ACTIVATED FACTOR XA HEAVY CHAIN, FACTOR X LIGHT CHAIN, ... | | Authors: | Groebke-Zbinden, K, Banner, D.W, Benz, J.M, Blasco, F, Decoret, G, Himber, J, Kuhn, B, Panday, N, Ricklin, F, Risch, P, Schlatter, D, Stahl, M, Unger, R, Haap, W. | | Deposit date: | 2008-06-26 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Design of Novel Aminopyrrolidine Factor Xa Inhibitors from a Screening Hit.
Eur.J.Med.Chem., 44, 2009
|
|
3C62
 
 | | Tetrameric Cytochrome cb562 (H59/D62/H63/H73/A74/H77) Assembly Stabilized by Interprotein Zinc Coordination | | Descriptor: | CALCIUM ION, PROTOPORPHYRIN IX CONTAINING FE, Soluble cytochrome b562, ... | | Authors: | Tezcan, F.A, Salgado, E.N, Lewis, R.A, Faraone-Mennella, J. | | Deposit date: | 2008-02-02 | | Release date: | 2008-05-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Metal-mediated self-assembly of protein superstructures: influence of secondary interactions on protein oligomerization and aggregation.
J.Am.Chem.Soc., 130, 2008
|
|
2VWW
 
 | | ephB4 kinase domain inhibitor complex | | Descriptor: | EPHRIN TYPE-B RECEPTOR 4, N'-(5-CHLORO-1,3-BENZODIOXOL-4-YL)-N-(3,4,5- TRIMETHOXYPHENYL)PYRIMIDINE-2,4-DIAMINE | | Authors: | Read, J, Brassington, C.A, Green, I, McCall, E.J, Valentine, A.L, Barratt, D, Leach, A.G, Kettle, J.G. | | Deposit date: | 2008-06-27 | | Release date: | 2008-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inhibitors of the Tyrosine Kinase Ephb4. Part 1: Structure-Based Design and Optimization of a Series of 2,4-Bis-Anilinopyrimidines
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3C7U
 
 | |
2VO1
 
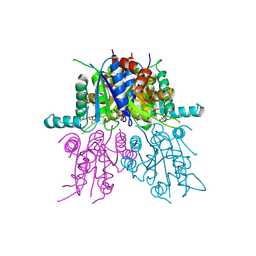 | | CRYSTAL STRUCTURE OF THE SYNTHETASE DOMAIN OF HUMAN CTP SYNTHETASE | | Descriptor: | CTP SYNTHASE 1, SULFATE ION | | Authors: | Stenmark, P, Kursula, P, Arrowsmith, C, Berglund, H, Edwards, A, Ehn, M, Flodin, S, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg-Schiavone, L, Kotenyoa, T, Moche, M, Nilsson-Ehle, P, Ogg, D, Persson, C, Sagemark, J, Schuler, H, Sundstrom, M, Thorsell, A.G, Van Den Berg, S, Weigelt, J, Nordlund, P. | | Deposit date: | 2008-02-08 | | Release date: | 2008-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Synthetase Domain of Human Ctp Synthetase, a Target for Anticancer Therapy.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
1ASN
 
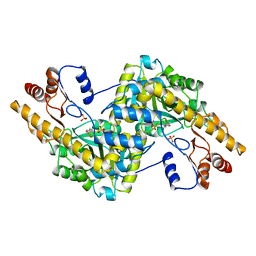 | |
3BY8
 
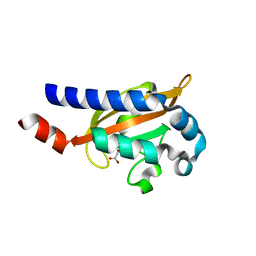 | | Crystal Structure of the E.coli DcuS Sensor Domain | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Sensor protein dcuS | | Authors: | Cheung, J, Hendrickson, W.A. | | Deposit date: | 2008-01-15 | | Release date: | 2008-08-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structures of C4-Dicarboxylate Ligand Complexes with Sensor Domains of Histidine Kinases DcuS and DctB.
J.Biol.Chem., 283, 2008
|
|
2VSP
 
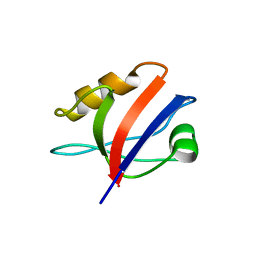 | | Crystal structure of the fourth PDZ domain of PDZ domain-containing protein 1 | | Descriptor: | PDZ DOMAIN-CONTAINING PROTEIN 1 | | Authors: | Yue, W.W, Shafqat, N, Pilka, E.S, Johansson, C, Murray, J.W, Elkins, J, Roos, A, Cooper, C, Phillips, C, Salah, E, von Delft, F, Doyle, D, Edwards, A, Wikstrom, M, Arrowsmith, C, Bountra, C, Oppermann, U. | | Deposit date: | 2008-04-28 | | Release date: | 2009-03-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal Structure of the Fourth Pdz Domain of Pdz Domain-Containing Protein 1
To be Published
|
|
2VVT
 
 | | Glutamate Racemase (MurI) from E. faecalis in complex with a 9-Benzyl Purine inhibitor | | Descriptor: | 2-butoxy-9-(2,6-difluorobenzyl)-N-(2-morpholin-4-ylethyl)-9H-purin-6-amine, D-GLUTAMIC ACID, GLUTAMATE RACEMASE | | Authors: | Geng, B, Breault, G, Comita-Prevoir, J, Petrichko, R, Eyermann, C, Lundqvist, T, Doig, P, Gorseth, E, Noonan, B. | | Deposit date: | 2008-06-11 | | Release date: | 2008-06-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Exploring 9-Benzyl Purines as Inhibitors of Glutamate Racemase (Muri) in Gram-Positive Bacteria.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2VUA
 
 | |
2VUP
 
 | |
3GMT
 
 | | Crystal structure of adenylate kinase from burkholderia pseudomallei | | Descriptor: | Adenylate kinase, SULFATE ION | | Authors: | Abendroth, J, Staker, B.L, Robinson, H, Buchko, G.W, Hewitt, S.N, Napuli, A.J, Van Voorhis, W, Stacy, R, Myler, P.J, Stewart, L, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2009-03-15 | | Release date: | 2009-06-02 | | Last modified: | 2013-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of Burkholderia pseudomallei adenylate kinase (Adk): profound asymmetry in the crystal structure of the 'open' state.
Biochem.Biophys.Res.Commun., 394, 2010
|
|
2W21
 
 | | Crystal structure of the aminoacid kinase domain of the glutamate 5 kinase of Escherichia coli. | | Descriptor: | GLUTAMATE 5-KINASE, SULFATE ION | | Authors: | Perez-Arellano, I, Gil-Ortiz, F, Marco-Marin, C, Cervera, J, Rubio, V. | | Deposit date: | 2008-10-21 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The Structure of Glutamate 5 Kinase of Escherichia Coli without Substrates
To be Published
|
|
5Z5M
 
 | |
3BU5
 
 | |
2VVC
 
 | | Aminopyrrolidine Factor Xa inhibitor | | Descriptor: | 5-chloro-N-[(3S,4S)-1-(2-{[2-fluoro-4-(2-oxopyridin-1(2H)-yl)phenyl]amino}-2-oxoethyl)-4-methoxypyrrolidin-3-yl]thiophene-2-carboxamide, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Groebke-Zbinden, K, Banner, D.W, Benz, J.M, Blasco, F, Decoret, G, Himber, J, Kuhn, B, Panday, N, Ricklin, F, Risch, P, Schlatter, D, Stahl, M, Unger, R, Haap, W. | | Deposit date: | 2008-06-05 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design of Novel Aminopyrrolidine Factor Xa Inhibitors from a Screening Hit.
Eur.J.Med.Chem., 44, 2009
|
|
3C2W
 
 | |
2W0S
 
 | | Crystal structure of vaccinia virus thymidylate kinase bound to brivudin-5'-monophosphate | | Descriptor: | (E)-5-(2-BROMOVINYL)-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Caillat, C, Topalis, D, Agrofoglio, L.A, Pochet, S, Balzarini, J, Deville-Bonne, D, Meyer, P. | | Deposit date: | 2008-10-08 | | Release date: | 2008-10-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.918 Å) | | Cite: | Crystal Structure of Poxvirus Thymidylate Kinase: An Unexpected Dimerization Has Implications for Antiviral Therapy
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3C63
 
 | | Tetrameric Cytochrome cb562 (K34/H59/D62/H63/H73/A74/H77) Assembly Stabilized by Interprotein Zinc Coordination | | Descriptor: | CALCIUM ION, PROTOPORPHYRIN IX CONTAINING FE, Soluble cytochrome b562, ... | | Authors: | Tezcan, F.A, Salgado, E.N, Lewis, R.A, Faraone-Mennella, J. | | Deposit date: | 2008-02-02 | | Release date: | 2008-05-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Metal-mediated self-assembly of protein superstructures: influence of secondary interactions on protein oligomerization and aggregation.
J.Am.Chem.Soc., 130, 2008
|
|
2W6R
 
 | | Crystal structure of an artificial (ba)8-barrel protein designed from identical half barrels | | Descriptor: | IMIDAZOLE GLYCEROL PHOSPHATE SYNTHASE SUBUNIT HISF | | Authors: | Hocker, B, Lochner, A, Seitz, T, Claren, J, Sterner, R. | | Deposit date: | 2008-12-18 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-Resolution Crystal Structure of an Artificial (Betaalpha)(8)-Barrel Protein Designed from Identical Half-Barrels.
Biochemistry, 48, 2009
|
|
2VO5
 
 | | Structural and biochemical evidence for a boat-like transition state in beta-mannosidases | | Descriptor: | (1R,4R,5R,7R,8R)-2-Benzyl-5-hydroxymethyl-2-aza-bicyclo[2.2.2]octane-4,7,8-triol, 1,2-ETHANEDIOL, BETA-MANNOSIDASE, ... | | Authors: | Tailford, L.E, Offen, W.A, Smith, N.L, Dumon, C, Moreland, C, Gratien, J, Heck, M.P, Stick, R.V, Bleriot, Y, Vasella, A, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2008-02-08 | | Release date: | 2008-04-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Biochemical Evidence for a Boat-Like Transition State in Beta-Mannosidases.
Nat.Chem.Biol., 4, 2008
|
|
