6EI6
 
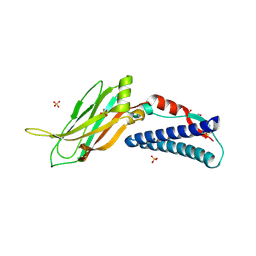 | | CC2D1B coordinates ESRCT-III activity during the mitotic reformation of the nuclear envelope | | Descriptor: | Coiled-coil and C2 domain-containing protein 1-like, DI(HYDROXYETHYL)ETHER, SULFATE ION | | Authors: | Ventimiglia, L.N, Cuesta-Geijo, M.A, Martinelli, N, Caballe, A, Macheboeuf, P, Miguet, N, Parnham, I.M, Olmos, Y, Carlton, J.G, Weissehorn, W, martin-Serrano, J. | | Deposit date: | 2017-09-18 | | Release date: | 2018-10-10 | | Last modified: | 2018-12-19 | | Method: | X-RAY DIFFRACTION (2.461 Å) | | Cite: | CC2D1B Coordinates ESCRT-III Activity during the Mitotic Reformation of the Nuclear Envelope.
Dev. Cell, 47, 2018
|
|
6ELY
 
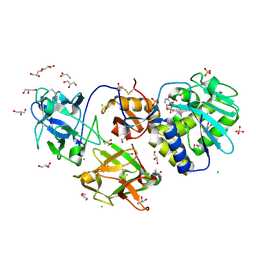 | | Crystal Structure of Mistletoe Lectin I (ML-I) from Viscum album in Complex with 4-N-Furfurylcytosine at 2.84 A Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-N-Furfurylcytosine, ... | | Authors: | Ahmad, M.S, Rasheed, S, Falke, S, Khaliq, B, Perbandt, M, Choudhary, M.I, Markiewicz, W.T, Barciszewski, J, Betzel, C. | | Deposit date: | 2017-09-30 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Crystal Structure of Mistletoe Lectin I (ML-I) from Viscum album in Complex with 4-N-Furfurylcytosine at 2.85 angstrom Resolution.
Med Chem, 14, 2018
|
|
5V30
 
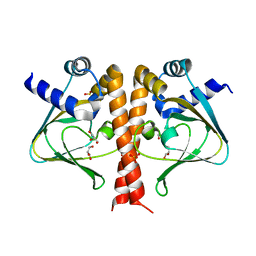 | | Crystal structure of the sensor domain of the transcriptional regulator HcpR from Porphyromonas Gingivalis | | Descriptor: | GLYCEROL, SULFATE ION, Transcriptional regulator | | Authors: | Musayev, F.N, Belvin, B.R, Escalante, C.R, Turner, J, Lewis, J.P. | | Deposit date: | 2017-03-06 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Nitrosative stress sensing in Porphyromonas gingivalis: structure of and heme binding by the transcriptional regulator HcpR.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6ELW
 
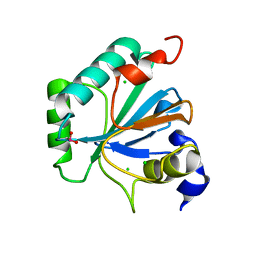 | | High resolution structure of selenocysteine containing human GPX4 | | Descriptor: | CHLORIDE ION, Phospholipid hydroperoxide glutathione peroxidase, mitochondrial | | Authors: | Kalms, J, Borchert, A, Kuhn, H, Scheerer, P. | | Deposit date: | 2017-09-29 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure and functional characterization of selenocysteine-containing glutathione peroxidase 4 suggests an alternative mechanism of peroxide reduction.
Biochim. Biophys. Acta, 1863, 2018
|
|
5V4L
 
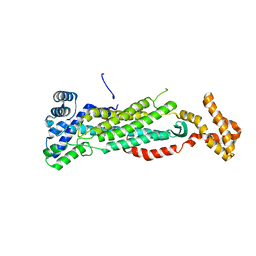 | |
5N9G
 
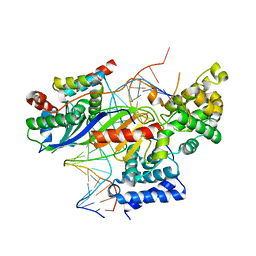 | | TFIIIB -TBP/Brf2/DNA and SANT domain of Bdp1- | | Descriptor: | DNA/RNA (25-MER), DNA/RNA (27-MER), SODIUM ION, ... | | Authors: | Gouge, J, Vannini, A, Guthertz, N. | | Deposit date: | 2017-02-24 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular mechanisms of Bdp1 in TFIIIB assembly and RNA polymerase III transcription initiation.
Nat Commun, 8, 2017
|
|
6EP7
 
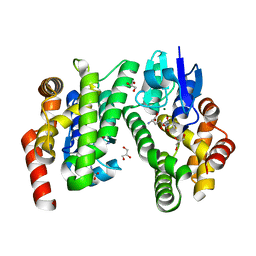 | | ARABIDOPSIS THALIANA GSTU23, GSH bound | | Descriptor: | GLUTATHIONE, GLYCEROL, Glutathione S-transferase U23, ... | | Authors: | Tossounian, M.A, Van Molle, I, Wahni, K, Jacques, S, Vertommen, D, Gevaert, K, Van Breusegem, F, Young, D, Rosado, L, Messens, J. | | Deposit date: | 2017-10-11 | | Release date: | 2018-04-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.947 Å) | | Cite: | Disulfide bond formation protects Arabidopsis thaliana glutathione transferase tau 23 from oxidative damage.
Biochim. Biophys. Acta, 1862, 2018
|
|
5UOJ
 
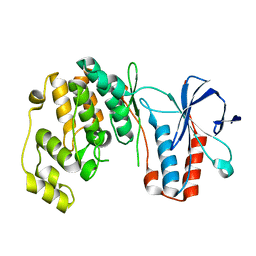 | |
5GZ2
 
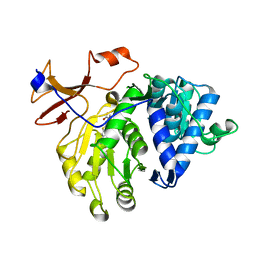 | | luciferase complex with 7-cy-L | | Descriptor: | (4S)-2-[6-(azepan-1-yl)-1,3-benzothiazol-2-yl]-4,5-dihydro-1,3-thiazole-4-carboxylic acid, Luciferin 4-monooxygenase | | Authors: | Su, J, Wang, F, Gu, L. | | Deposit date: | 2016-09-26 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | luciferase complex with 7-cy-L
To Be Published
|
|
5GZA
 
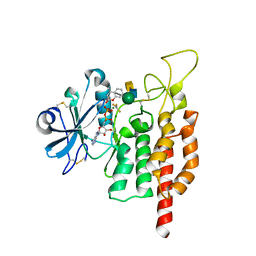 | | protein O-mannose kinase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-mannopyranose, 4-METHYL-2H-CHROMEN-2-ONE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Xiao, J. | | Deposit date: | 2016-09-27 | | Release date: | 2016-12-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of protein O-mannose kinase reveals a unique active site architecture
Elife, 5, 2016
|
|
5H3B
 
 | |
5H3O
 
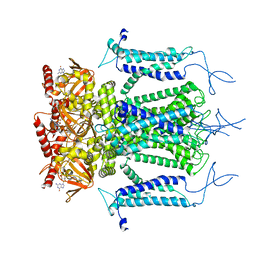 | | Structure of a eukaryotic cyclic nucleotide-gated channel | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, Cyclic nucleotide-gated cation channel, SODIUM ION | | Authors: | Li, M, Zhou, X, Wang, S, Michailidis, I, Gong, Y, Su, D, Li, H, Li, X, Yang, J. | | Deposit date: | 2016-10-26 | | Release date: | 2017-01-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of a eukaryotic cyclic-nucleotide-gated channel.
Nature, 542, 2017
|
|
1H6N
 
 | | Formation of a tyrosyl radical intermediate in Proteus mirabilis catalase by directed mutagenesis and consequences for nucleotide reactivity | | Descriptor: | ACETATE ION, CATALASE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Andreoletti, P, Sainz, G, Jaquinod, M, Gagnon, J, Jouve, H.M. | | Deposit date: | 2001-06-20 | | Release date: | 2003-10-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | High Resolution Structure and Biochemical Properties of a Recombinant Proteus Mirabilis Catalase Depleted in Iron.
Proteins: Struct.,Funct., Genet., 50, 2003
|
|
5MZZ
 
 | | Crystal structure of the decarboxylase AibA/AibB in complex with 3-methylglutaconate | | Descriptor: | 3-methylpent-2-enedioic acid, ACETATE ION, GLYCEROL, ... | | Authors: | Bock, T, Luxenburger, E, Hoffmann, J, Schuetza, V, Feiler, C, Mueller, R, Blankenfeldt, W. | | Deposit date: | 2017-02-02 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | AibA/AibB Induces an Intramolecular Decarboxylation in Isovalerate Biosynthesis by Myxococcus xanthus.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5FMV
 
 | | Crystal structure of human CD45 extracellular region, domains d1-d4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, RECEPTOR-TYPE TYROSINE-PROTEIN PHOSPHATASE C, SULFATE ION | | Authors: | Chang, V.T, Fernandes, R.A, Ganzinger, K.A, Lee, S.F, Siebold, C, McColl, J, Jonsson, P, Palayret, M, Harlos, K, Coles, C.H, Jones, E.Y, Lui, Y, Huang, E, Gilbert, R.J.C, Klenerman, D, Aricescu, A.R, Davis, S.J. | | Deposit date: | 2015-11-09 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Initiation of T Cell Signaling by Cd45 Segregation at 'Close Contacts'.
Nat.Immunol., 17, 2016
|
|
5FKC
 
 | |
5FN6
 
 | | Crystal structure of human CD45 extracellular region, domains d1-d3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, RECEPTOR-TYPE TYROSINE-PROTEIN PHOSPHATASE C | | Authors: | Chang, V.T, Fernandes, R.A, Ganzinger, K.A, Lee, S.F, Siebold, C, McColl, J, Jonsson, P, Palayret, M, Harlos, K, Coles, C.H, Jones, E.Y, Lui, Y, Huang, E, Gilbert, R.J.C, Klenerman, D, Aricescu, A.R, Davis, S.J. | | Deposit date: | 2015-11-10 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Initiation of T Cell Signaling by Cd45 Segregation at 'Close Contacts'.
Nat.Immunol., 17, 2016
|
|
5FKV
 
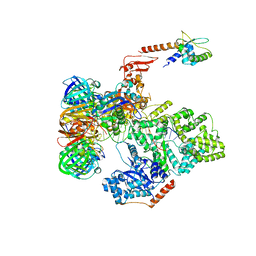 | | cryo-EM structure of the E. coli replicative DNA polymerase complex bound to DNA (DNA polymerase III alpha, beta, epsilon, tau complex) | | Descriptor: | DNA POLYMERASE III BETA, DNA POLYMERASE III EPSILON, DNA POLYMERASE III SUBUNIT ALPHA, ... | | Authors: | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | Deposit date: | 2015-10-20 | | Release date: | 2015-11-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.04 Å) | | Cite: | cryo-EM structures of theE. colireplicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease andtau.
Elife, 4, 2015
|
|
5FN8
 
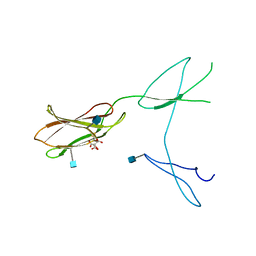 | | Crystal structure of rat CD45 extracellular region, domains d3-d4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRATE ANION, RECEPTOR-TYPE TYROSINE-PROTEIN PHOSPHATASE C | | Authors: | Chang, V.T, Fernandes, R.A, Ganzinger, K.A, Lee, S.F, Siebold, C, McColl, J, Jonsson, P, Palayret, M, Harlos, K, Coles, C.H, Jones, E.Y, Lui, Y, Huang, E, Gilbert, R.J.C, Klenerman, D, Aricescu, A.R, Davis, S.J. | | Deposit date: | 2015-11-11 | | Release date: | 2016-03-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Initiation of T Cell Signaling by Cd45 Segregation at 'Close Contacts'.
Nat.Immunol., 17, 2016
|
|
5N13
 
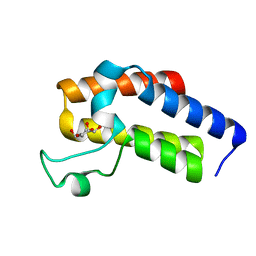 | | Second Bromodomain (BD2) from Candida albicans Bdf1 in the unbound form | | Descriptor: | Bromodomain-containing factor 1, GLYCEROL | | Authors: | Mietton, F, Ferri, E, Champlebouxm, M, Zala, N, Maubon, D, Zhou, Y, Harbut, M, Spittler, D, Garnaud, C, Courcon, M, Chauvel, M, d'Enfert, C, Kashemirov, B.A, Hull, M, Cornet, M, McKenna, C.E, Govin, J, Petosa, C. | | Deposit date: | 2017-02-04 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Selective BET bromodomain inhibition as an antifungal therapeutic strategy.
Nat Commun, 8, 2017
|
|
5FNC
 
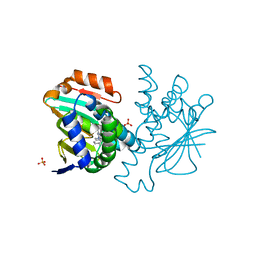 | | Dynamic Undocking and the Quasi-Bound State as tools for Drug Design | | Descriptor: | 6-CHLORO-4-N-[(4-METHYLPHENYL)METHYL]PYRIMIDINE- 2,4-DIAMINE, HEAT SHOCK PROTEIN, HSP90-ALPHA, ... | | Authors: | Ruiz-Carmona, S, Schmidtke, P, Luque, F.J, Baker, L.M, Matassova, N, Davis, B, Roughley, S, Murray, J, Hubbard, R, Barril, X. | | Deposit date: | 2015-11-13 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dynamic undocking and the quasi-bound state as tools for drug discovery.
Nat Chem, 9, 2017
|
|
5N17
 
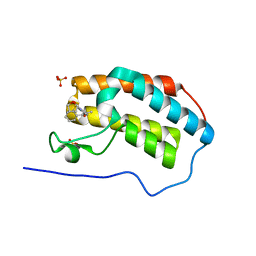 | | First Bromodomain (BD1) from Candida albicans Bdf1 bound to a dibenzothiazepinone (compound 3) | | Descriptor: | (2~{S})-~{N}-(5-methyl-6-oxidanylidene-benzo[b][1,4]benzothiazepin-2-yl)oxolane-2-carboxamide, Bromodomain-containing factor 1, SULFATE ION | | Authors: | Mietton, F, Ferri, E, Champleboux, M, Zala, N, Maubon, D, Zhou, Y, Harbut, M, Spittler, D, Garnaud, C, Courcon, M, Chauvon, M, d'Enfer, C, Kashemirov, B.A, Hull, M, Cornet, M, McKenna, C.E, Govin, J, Petosa, C. | | Deposit date: | 2017-02-05 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Selective BET bromodomain inhibition as an antifungal therapeutic strategy.
Nat Commun, 8, 2017
|
|
5FTX
 
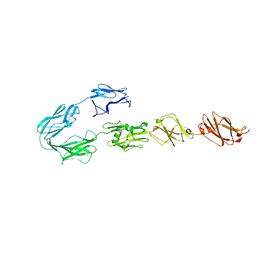 | | Structure of surface layer protein SbsC, domains 4-9 | | Descriptor: | CALCIUM ION, SURFACE LAYER PROTEIN, ZINC ION | | Authors: | Dordic, A, Pavkov-Keller, T, Eder, M, Egelseer, E.M, Davis, K, Mills, D, Sleytr, U.B, Kuehlbrandt, W, Vonck, J, Keller, W. | | Deposit date: | 2016-01-18 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structure of Surface Layer Protein Sbsc
To be Published
|
|
6EM5
 
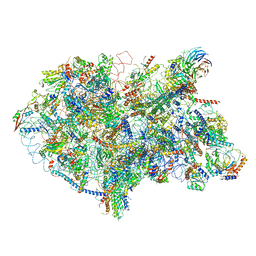 | | State D architectural model (Nsa1-TAP Flag-Ytm1) - Visualizing the assembly pathway of nucleolar pre-60S ribosomes | | Descriptor: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 25S ribosomal RNA, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | Authors: | Kater, L, Cheng, J, Barrio-Garcia, C, Hurt, E, Beckmann, R. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Visualizing the Assembly Pathway of Nucleolar Pre-60S Ribosomes.
Cell, 171, 2017
|
|
6EV1
 
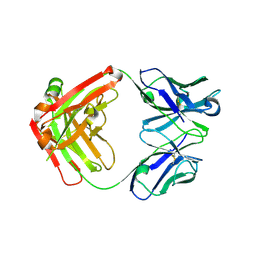 | | Crystal structure of antibody against schizophyllan | | Descriptor: | Heavy chain, Light chain | | Authors: | Sung, K.H, Josewski, J, Dubel, S, Blankenfeldt, W, Rau, U. | | Deposit date: | 2017-11-01 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.043 Å) | | Cite: | Structural insights into antigen recognition of an anti-beta-(1,6)-beta-(1,3)-D-glucan antibody.
Sci Rep, 8, 2018
|
|
