5SM5
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z2856434807 | | Descriptor: | 2-[4-(2-methoxyphenyl)piperazin-1-yl]ethanenitrile, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SLB
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z744930860 | | Descriptor: | 3-methyl-N-(2-methylbutan-2-yl)-1H-pyrazole-5-carboxamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SLW
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z1310876699 | | Descriptor: | 2-fluoro-N-[2-(pyridin-4-yl)ethyl]benzamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SLK
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z1354370680 | | Descriptor: | 2-[(5-chloro-3-fluoropyridin-2-yl)(methyl)amino]ethan-1-ol, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SLS
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z1373445602 | | Descriptor: | 4-(3-fluoranylpyridin-2-yl)-1-methyl-piperazin-2-one, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SL1
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z1273312153 | | Descriptor: | N-methyl-1H-indole-7-carboxamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.383 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SM1
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z68277692 | | Descriptor: | N-methyl-N-[2-(pyridin-2-yl)ethyl]benzamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.941 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SMG
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z2092370954 | | Descriptor: | 3-amino-N-ethyl-N-methylbenzamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SL2
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z100643660 | | Descriptor: | N,1-dimethyl-1H-indole-3-carboxamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.739 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SLU
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z1796014543 | | Descriptor: | 1-[(2-fluorophenyl)methyl]-N-methylcyclopropane-1-carboxamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SMA
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z2856434890 | | Descriptor: | 4-methyl-N-phenylpiperazine-1-carboxamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SKW
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z1272494722 | | Descriptor: | PHOSPHATE ION, Proofreading exoribonuclease nsp14, ZINC ION, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SLV
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z2856434942 | | Descriptor: | PHOSPHATE ION, Proofreading exoribonuclease nsp14, ZINC ION, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SL7
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z1186029914 | | Descriptor: | (3R)-1-(2-fluorophenyl)-3-(methylamino)pyrrolidin-2-one, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.841 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SMH
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z2856434938 | | Descriptor: | 1-(5-methoxy-1H-indol-3-yl)-N,N-dimethyl-methanamine, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5RY4
 
 | | INPP5D PanDDA analysis group deposition -- Crystal Structure of the phosphatase and C2 domains of SHIP1 in complex with Z300245038 | | Descriptor: | DIMETHYL SULFOXIDE, N-[(3-fluorophenyl)methyl]-N-methylsulfuric diamide, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
5RZ9
 
 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z943693514 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Isoform 2 of Band 4.1-like protein 3, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
5RZP
 
 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z33546965 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Isoform 2 of Band 4.1-like protein 3, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
5RWY
 
 | | INPP5D PanDDA analysis group deposition -- Crystal Structure of the phosphatase and C2 domains of SHIP1 in complex with Z1259335913 | | Descriptor: | 1-{1-[(2-methyl-1,3-thiazol-4-yl)methyl]piperidin-4-yl}methanamine, DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
5RYU
 
 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z1545196403 | | Descriptor: | 1,2-ETHANEDIOL, 2-fluoro-N-[(1H-pyrazol-3-yl)methyl]aniline, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
5RZ3
 
 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z1259341037 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Isoform 2 of Band 4.1-like protein 3, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
5RZY
 
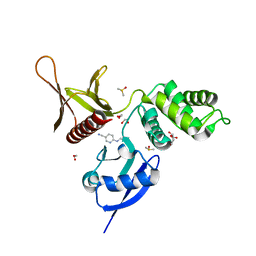 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z219104216 | | Descriptor: | 1,2-ETHANEDIOL, 6-(ethylamino)pyridine-3-carbonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
5RZV
 
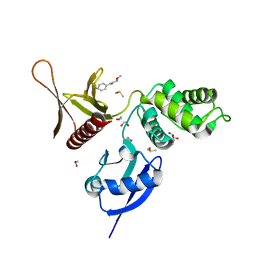 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z31504642 | | Descriptor: | 1,2-ETHANEDIOL, 2-methoxy-N-phenylacetamide, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
5RW5
 
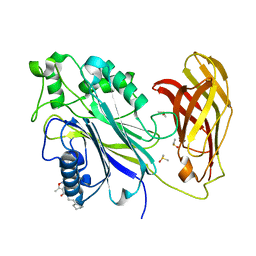 | | INPP5D PanDDA analysis group deposition -- Crystal Structure of the phosphatase and C2 domains of SHIP1 in complex with Z69092635 | | Descriptor: | DIMETHYL SULFOXIDE, N-(2-cyclohexylethyl)-3,5-dimethyl-1,2-oxazole-4-carboxamide, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
5RYB
 
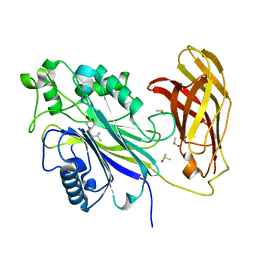 | | INPP5D PanDDA analysis group deposition -- Crystal Structure of the phosphatase and C2 domains of SHIP1 in complex with Z927400026 | | Descriptor: | (2S)-2-amino-3-methyl-1-(morpholin-4-yl)butan-1-one, DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
