7QD8
 
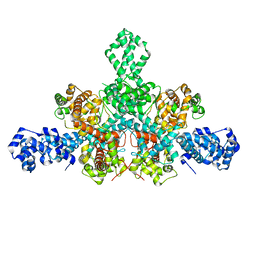 | | Cryo-EM structure of Tn4430 TnpA transposase from Tn3 family in apo state | | Descriptor: | Transposase for transposon Tn4430 | | Authors: | Shkumatov, A.V, Oger, C.A, Aryanpour, N, Hallet, B.F, Efremov, R.G. | | Deposit date: | 2021-11-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insight into Tn3 family transposition mechanism.
Nat Commun, 13, 2022
|
|
7QD4
 
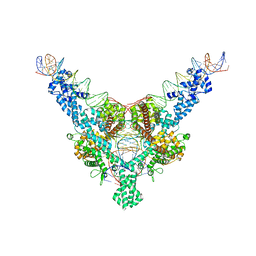 | | Cryo-EM structure of Tn4430 TnpA transposase from Tn3 family in complex with 100 bp long transposon end DNA | | Descriptor: | IR100 DNA substrate, none transferred strand, transferred strand, ... | | Authors: | Shkumatov, A.V, Oger, C.A, Aryanpour, N, Hallet, B.F, Efremov, R.G. | | Deposit date: | 2021-11-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insight into Tn3 family transposition mechanism.
Nat Commun, 13, 2022
|
|
7QD6
 
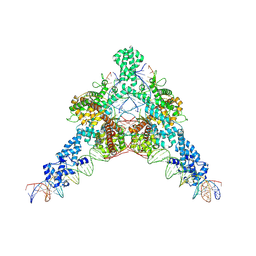 | | Cryo-EM structure of Tn4430 TnpA transposase from Tn3 family in complex with strand-transfer like DNA product | | Descriptor: | IR71st non transferred strand, IR71st transferred strand, Transposase for transposon Tn4430 | | Authors: | Shkumatov, A.V, Oger, C.A, Aryanpour, N, Hallet, B.F, Efremov, R.G. | | Deposit date: | 2021-11-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insight into Tn3 family transposition mechanism.
Nat Commun, 13, 2022
|
|
7QD5
 
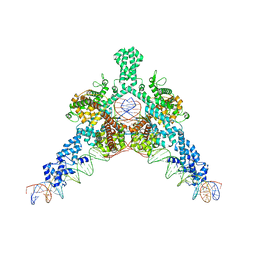 | | Cryo-EM structure of Tn4430 TnpA transposase from Tn3 family in complex with 48 bp long transposon end DNA | | Descriptor: | IR48 DNA substrate, non transferred strand, IR48 transferred strand, ... | | Authors: | Shkumatov, A.V, Oger, C.A, Aryanpour, N, Hallet, B.F, Efremov, R.G. | | Deposit date: | 2021-11-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insight into Tn3 family transposition mechanism.
Nat Commun, 13, 2022
|
|
7NZ1
 
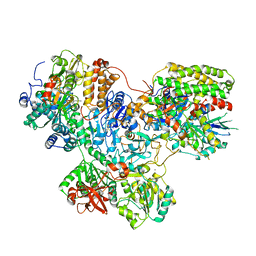 | |
7NYR
 
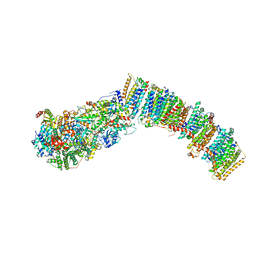 | |
7NYU
 
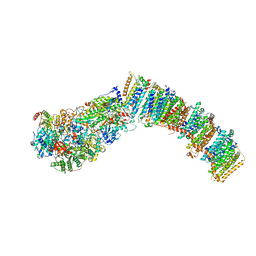 | |
7NYV
 
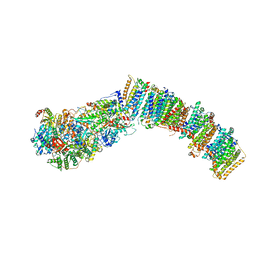 | |
7Z09
 
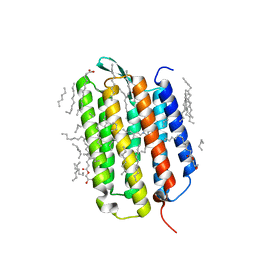 | | Crystal structure of the ground state of bacteriorhodopsin at 1.05 Angstrom resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin, EICOSANE, ... | | Authors: | Borshchevskiy, V, Kovalev, K, Round, E, Efremov, R, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-02-22 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | True-atomic-resolution insights into the structure and functional role of linear chains and low-barrier hydrogen bonds in proteins.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7Z0A
 
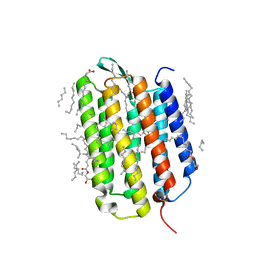 | | Crystal structure of the ground state of bacteriorhodopsin at 1.22 Angstrom resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin, EICOSANE, ... | | Authors: | Borshchevskiy, V, Kovalev, K, Round, E, Efremov, R, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-02-22 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | True-atomic-resolution insights into the structure and functional role of linear chains and low-barrier hydrogen bonds in proteins.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7Z0D
 
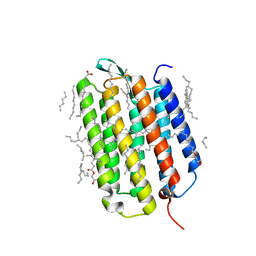 | | Crystal structure of the L state of bacteriorhodopsin at 1.20 Angstrom resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin, EICOSANE, ... | | Authors: | Borshchevskiy, V, Kovalev, K, Round, E, Efremov, R, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-02-22 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | True-atomic-resolution insights into the structure and functional role of linear chains and low-barrier hydrogen bonds in proteins.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7Z0C
 
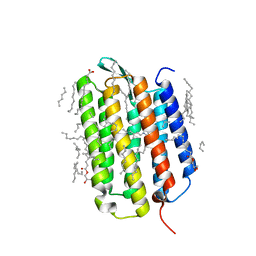 | | Crystal structure of the K state of bacteriorhodopsin at 1.53 Angstrom resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin, EICOSANE, ... | | Authors: | Borshchevskiy, V, Kovalev, K, Round, E, Efremov, R, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-02-22 | | Release date: | 2022-05-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | True-atomic-resolution insights into the structure and functional role of linear chains and low-barrier hydrogen bonds in proteins.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7Z0E
 
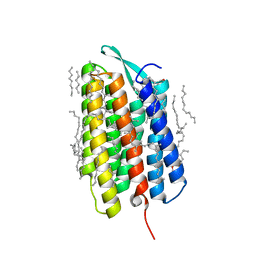 | | Crystal structure of the M state of bacteriorhodopsin at 1.22 Angstrom resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (6E,10E,14E,18E)-2,6,10,15,19,23-hexamethyltetracosa-2,6,10,14,18,22-hexaene, 2,3-DI-PHYTANYL-GLYCEROL, ... | | Authors: | Borshchevskiy, V, Kovalev, K, Round, E, Efremov, R, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-02-22 | | Release date: | 2022-05-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | True-atomic-resolution insights into the structure and functional role of linear chains and low-barrier hydrogen bonds in proteins.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7NYH
 
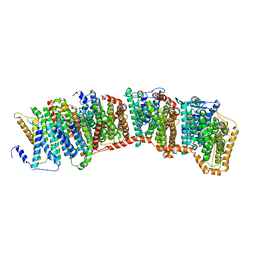 | |
8BM0
 
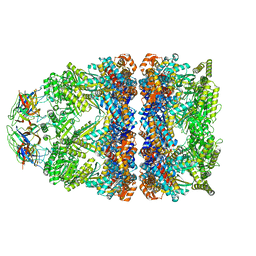 | | Structure of GroEL:GroES-ATP complex plunge frozen 200 ms after reaction initiation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperonin GroEL, Co-chaperonin GroES, ... | | Authors: | Dhurandhar, M, Torino, S, Efremov, R. | | Deposit date: | 2022-11-10 | | Release date: | 2023-08-09 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Time-resolved cryo-EM using a combination of droplet microfluidics with on-demand jetting.
Nat.Methods, 20, 2023
|
|
8BK9
 
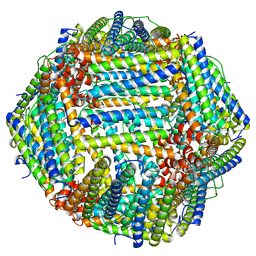 | |
8BKB
 
 | |
8BMO
 
 | | Structure of GroEL:GroES complex exhibiting ADP-conformation in trans ring obtained under the continuous turnover conditions | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperonin GroEL, Co-chaperonin GroES, ... | | Authors: | Dhurandhar, M, Torino, S, Efremov, R. | | Deposit date: | 2022-11-10 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Time-resolved cryo-EM using a combination of droplet microfluidics with on-demand jetting.
Nat.Methods, 20, 2023
|
|
8BKA
 
 | |
8BKZ
 
 | | GroEL:GroES-ATP complex under continuous turnover conditions | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperonin GroEL, Co-chaperonin GroES, ... | | Authors: | Dhurandhar, M, Torino, S, Efremov, R. | | Deposit date: | 2022-11-09 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Time-resolved cryo-EM using a combination of droplet microfluidics with on-demand jetting.
Nat.Methods, 20, 2023
|
|
8BM1
 
 | | Structure of GroEL:GroES-ATP complex under continuous turnover conditions | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperonin GroEL, Co-chaperonin GroES, ... | | Authors: | Dhurandhar, M, Torino, S, Efremov, R. | | Deposit date: | 2022-11-10 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Time-resolved cryo-EM using a combination of droplet microfluidics with on-demand jetting.
Nat.Methods, 20, 2023
|
|
8BMT
 
 | | Structure of GroEL:GroES-ATP complex plunge frozen 200 ms after reaction initiation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperonin GroEL, Co-chaperonin GroES, ... | | Authors: | Dhurandhar, M, Efremov, R, Torino, S. | | Deposit date: | 2022-11-10 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Time-resolved cryo-EM using a combination of droplet microfluidics with on-demand jetting.
Nat.Methods, 20, 2023
|
|
8BL7
 
 | |
8BLD
 
 | |
8BLY
 
 | |
