1QL6
 
 | | THE CATALYTIC MECHANISM OF PHOSPHORYLASE KINASE PROBED BY MUTATIONAL STUDIES | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, PHOSPHORYLASE KINASE, ... | | Authors: | Skamnaki, V.T, Owen, D.J, Noble, M.E.M, Lowe, E.D, Oikonomakos, N.G, Johnson, L.N. | | Deposit date: | 1999-08-24 | | Release date: | 1999-12-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Catalytic Mechanism of Phosphorylase Kinase Probed by Mutational Studies.
Biochemistry, 38, 1999
|
|
1OIY
 
 | | Structure of human Thr160-phospho CDK2/cyclin A complexed with a 6-cyclohexylmethyloxy-2-anilino-purine inhibitor | | Descriptor: | 4-(6-CYCLOHEXYLMETHOXY-9H-PURIN-2-YLAMINO)--BENZAMIDE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, ... | | Authors: | Pratt, D.J, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2003-06-26 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | N2-Substituted O6-Cyclohexylmethylguanine Derivatives: Potent Inhibitors of Cyclin-Dependent Kinases 1 and 2
J.Med.Chem., 47, 2004
|
|
1OI9
 
 | | Structure of human Thr160-phospho CDK2/cyclin A complexed with a 6-cyclohexylmethyloxy-2-anilino-purine inhibitor | | Descriptor: | 6-CYCLOHEXYLMETHYLOXY-2-(4'-HYDROXYANILINO)PURINE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, ... | | Authors: | Pratt, D.J, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2003-06-10 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | N2-Substituted O6-Cyclohexylmethylguanine Derivatives: Potent Inhibitors of Cyclin-Dependent Kinases 1 and 2
J.Med.Chem., 47, 2004
|
|
1OIU
 
 | | Structure of human Thr160-phospho CDK2/cyclin A complexed with a 6-cyclohexylmethyloxy-2-anilino-purine inhibitor | | Descriptor: | 3-(6-CYCLOHEXYLMETHOXY-9H-PURIN-2-YLAMINO)-BENZENESULFONAMIDE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, ... | | Authors: | Pratt, D.J, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2003-06-26 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | N2-Substituted O6-Cyclohexylmethylguanine Derivatives: Potent Inhibitors of Cyclin-Dependent Kinases 1 and 2
J.Med.Chem., 47, 2004
|
|
1QHJ
 
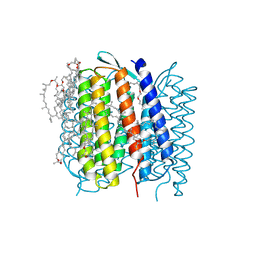 | | X-RAY STRUCTURE OF BACTERIORHODOPSIN GROWN IN LIPIDIC CUBIC PHASES | | Descriptor: | 1,2-[DI-2,6,10,14-TETRAMETHYL-HEXADECAN-16-OXY]-PROPANE, PROTEIN (BACTERIORHODOPSIN), RETINAL | | Authors: | Belrhali, H, Nollert, P, Royant, A, Menzel, C, Rosenbusch, J.P, Landau, E.M, Pebay-Peyroula, E. | | Deposit date: | 1999-05-04 | | Release date: | 1999-07-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein, lipid and water organization in bacteriorhodopsin crystals: a molecular view of the purple membrane at 1.9 A resolution.
Structure Fold.Des., 7, 1999
|
|
2N9O
 
 | | Solution structure of RNF126 N-terminal zinc finger domain | | Descriptor: | E3 ubiquitin-protein ligase RNF126, ZINC ION | | Authors: | Martinez-Lumbreras, S, Krysztofinska, E.M, Thapaliya, A, Isaacson, R.L. | | Deposit date: | 2015-12-01 | | Release date: | 2016-05-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and functional insights into the E3 ligase, RNF126.
Sci Rep, 6, 2016
|
|
5MSJ
 
 | | Mouse PA28alpha | | Descriptor: | Proteasome activator complex subunit 1 | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2017-01-05 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The Mammalian Proteasome Activator PA28 Forms an Asymmetric alpha 4 beta 3 Complex.
Structure, 25, 2017
|
|
1QKO
 
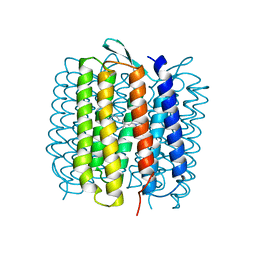 | | HIGH RESOLUTION X-RAY STRUCTURE OF AN EARLY INTERMEDIATE IN THE BACTERIORHODOPSIN PHOTOCYCLE | | Descriptor: | BACTERIORHODOPSIN, RETINAL | | Authors: | Edman, K, Nollert, P, Royant, A, Belrhali, H, Pebay-Peyroula, E, Hajdu, J, Neutze, R, Landau, E.M. | | Deposit date: | 1999-07-30 | | Release date: | 1999-10-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High Resolution X-Ray Structure of an Early Intermediate in the Bacteriorhodopsin Photocycle
Nature, 401, 1999
|
|
5N1T
 
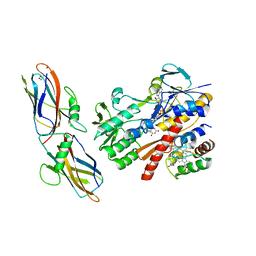 | | Crystal structure of complex between flavocytochrome c and copper chaperone CopC from T. paradoxus | | Descriptor: | COPPER (II) ION, CopC, Cytochrome C, ... | | Authors: | Osipov, E.M, Lilina, A.V, Tikhonova, T.V, Tsallagov, S.I, Popov, V.O. | | Deposit date: | 2017-02-06 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the flavocytochrome c sulfide dehydrogenase associated with the copper-binding protein CopC from the haloalkaliphilic sulfur-oxidizing bacterium Thioalkalivibrio paradoxusARh 1.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5N53
 
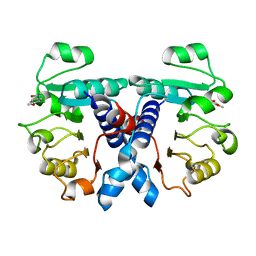 | | Crystal structure of human 3-phosphoglycerate dehydrogenase in complex with N-(3-chloro-4-methoxyphenyl) acetamide | | Descriptor: | D-3-phosphoglycerate dehydrogenase, ~{N}-(3-chloranyl-4-methoxy-phenyl)ethanamide | | Authors: | Unterlass, J.E, Basle, A, Blackburn, T.J, Tucker, J, Cano, C, Noble, M.E.M, Curtin, N.J. | | Deposit date: | 2017-02-12 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Validating and enabling phosphoglycerate dehydrogenase (PHGDH) as a target for fragment-based drug discovery in PHGDH-amplified breast cancer.
Oncotarget, 9, 2018
|
|
5NJB
 
 | | E. coli Microcin-processing metalloprotease TldD/E with actinonin bound | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACTINONIN, ... | | Authors: | Ghilarov, D, Serebryakova, M, Stevenson, C.E.M, Hearnshaw, S.J, Volkov, D, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2017-03-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Origins of Specificity in the Microcin-Processing Protease TldD/E.
Structure, 25, 2017
|
|
5NJF
 
 | | E. coli Microcin-processing metalloprotease TldD/E (TldD H262A mutant) with pentapeptide bound | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ALA-ALA-ALA-ALA-ALA, ... | | Authors: | Ghilarov, D, Serebryakova, M, Stevenson, C.E.M, Hearnshaw, S.J, Volkov, D, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2017-03-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | The Origins of Specificity in the Microcin-Processing Protease TldD/E.
Structure, 25, 2017
|
|
5CT9
 
 | | G158E/K44E/R57E/Y49E Bacillus subtilis lipase A with 5% [BMIM][Cl] | | Descriptor: | 1-butyl-3-methyl-1H-imidazol-3-ium, CHLORIDE ION, Esterase | | Authors: | Nordwald, E.M, Plaks, J.G, Snell, J.R, Sousa, M.C, Kaar, J.L. | | Deposit date: | 2015-07-23 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystallographic Investigation of Imidazolium Ionic Liquid Effects on Enzyme Structure.
Chembiochem, 16, 2015
|
|
5CPS
 
 | | Disproportionating enzyme 1 from Arabidopsis - maltotriose soak | | Descriptor: | 1,2-ETHANEDIOL, 4-alpha-glucanotransferase DPE1, chloroplastic/amyloplastic, ... | | Authors: | O'Neill, E.C, Stevenson, C.E.M, Tantanarat, K, Latousakis, D, Donaldson, M.I, Rejzek, M, Limpaseni, T, Smith, A.M, Field, R.A, Lawson, D.M. | | Deposit date: | 2015-07-21 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Dissection of the Maltodextrin Disproportionation Cycle of the Arabidopsis Plastidial Disproportionating Enzyme 1 (DPE1).
J.Biol.Chem., 290, 2015
|
|
5LWZ
 
 | | Cys-Gly dipeptidase GliJ (space group C2) | | Descriptor: | Dipeptidase, FE (III) ION, GLYCEROL | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2016-09-19 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
ACS Chem. Biol., 12, 2017
|
|
5CRI
 
 | | Wild-type Bacillus subtilis lipase A with 0% [BMIM][Cl] | | Descriptor: | Esterase, SULFATE ION | | Authors: | Nordwald, E.M, Plaks, J.G, Snell, J.R, Sousa, M.C, Kaar, J.L. | | Deposit date: | 2015-07-22 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystallographic Investigation of Imidazolium Ionic Liquid Effects on Enzyme Structure.
Chembiochem, 16, 2015
|
|
5CT6
 
 | | Wild-type Bacillus subtilis lipase A with 20% [BMIM][Cl] | | Descriptor: | 1-butyl-3-methyl-1H-imidazol-3-ium, CHLORIDE ION, Lipase EstA | | Authors: | Nordwald, E.M, Plaks, J.G, Snell, J.R, Sousa, M.C, Kaar, J.L. | | Deposit date: | 2015-07-23 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Crystallographic Investigation of Imidazolium Ionic Liquid Effects on Enzyme Structure.
Chembiochem, 16, 2015
|
|
5CT8
 
 | | G158E/K44E/R57E/Y49E Bacillus subtilis lipase A with 0% [BMIM][Cl] | | Descriptor: | Quadruple mutant lipase A, SULFATE ION | | Authors: | Nordwald, E.M, Plaks, J.G, Snell, J.R, Sousa, M.C, Kaar, J.L. | | Deposit date: | 2015-07-23 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Crystallographic Investigation of Imidazolium Ionic Liquid Effects on Enzyme Structure.
Chembiochem, 16, 2015
|
|
5CPQ
 
 | | Disproportionating enzyme 1 from Arabidopsis - apo form | | Descriptor: | 1,2-ETHANEDIOL, 4-alpha-glucanotransferase DPE1, chloroplastic/amyloplastic | | Authors: | O'Neill, E.C, Stevenson, C.E.M, Tantanarat, K, Latousakis, D, Donaldson, M.I, Rejzek, M, Limpaseni, T, Smith, A.M, Field, R.A, Lawson, D.M. | | Deposit date: | 2015-07-21 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural Dissection of the Maltodextrin Disproportionation Cycle of the Arabidopsis Plastidial Disproportionating Enzyme 1 (DPE1).
J.Biol.Chem., 290, 2015
|
|
5CT4
 
 | | Wild-type Bacillus subtilis lipase A with 5% [BMIM][Cl] | | Descriptor: | 1-butyl-3-methyl-1H-imidazol-3-ium, CHLORIDE ION, Esterase, ... | | Authors: | Nordwald, E.M, Plaks, J.G, Snell, J.R, Sousa, M.C, Kaar, J.L. | | Deposit date: | 2015-07-23 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystallographic Investigation of Imidazolium Ionic Liquid Effects on Enzyme Structure.
Chembiochem, 16, 2015
|
|
5CQ1
 
 | | Disproportionating enzyme 1 from Arabidopsis - cycloamylose soak | | Descriptor: | 1,2-ETHANEDIOL, 4-alpha-glucanotransferase DPE1, chloroplastic/amyloplastic, ... | | Authors: | O'Neill, E.C, Stevenson, C.E.M, Tantanarat, K, Latousakis, D, Donaldson, M.I, Rejzek, M, Limpaseni, T, Smith, A.M, Field, R.A, Lawson, D.M. | | Deposit date: | 2015-07-21 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Dissection of the Maltodextrin Disproportionation Cycle of the Arabidopsis Plastidial Disproportionating Enzyme 1 (DPE1).
J.Biol.Chem., 290, 2015
|
|
5LX4
 
 | | Cys-Gly dipeptidase GliJ mutant D38H | | Descriptor: | Dipeptidase, FE (III) ION, GLYCEROL | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2016-09-20 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
ACS Chem. Biol., 12, 2017
|
|
5CZA
 
 | | Yeast 20S proteasome beta5-D166N mutant | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-07-31 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A unified mechanism for proteolysis and autocatalytic activation in the 20S proteasome.
Nat Commun, 7, 2016
|
|
5NEV
 
 | | CDK2/Cyclin A in complex with compound 73 | | Descriptor: | 4-[[6-(3-phenylphenyl)-7~{H}-purin-2-yl]amino]benzenesulfonamide, Cyclin-A2, Cyclin-dependent kinase 2 | | Authors: | Coxon, C.R, Anscombe, E, Harnor, S.J, Martin, M.P, Carbain, B, Hardcastle, I.R, Harlow, L.K, Korolchuk, S, Matheson, C.J, Noble, M.E.M, Newell, D.R, Turner, D, Sivaprakasam, M, Wang, L.Z, Wong, C, Golding, B.T, Griffin, R.J, Cano, G. | | Deposit date: | 2017-03-12 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Cyclin-Dependent Kinase (CDK) Inhibitors: Structure-Activity Relationships and Insights into the CDK-2 Selectivity of 6-Substituted 2-Arylaminopurines.
J. Med. Chem., 60, 2017
|
|
1QA7
 
 | | CRYSTAL COMPLEX OF THE 3C PROTEINASE FROM HEPATITIS A VIRUS WITH ITS INHIBITOR AND IMPLICATIONS FOR THE POLYPROTEIN PROCESSING IN HAV | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, HAV 3C PROTEINASE, ... | | Authors: | Bergmann, E.M, Cherney, M.M, Mckendrick, J, Vederas, J.C, James, M.N.G. | | Deposit date: | 1999-04-15 | | Release date: | 1999-04-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an inhibitor complex of the 3C proteinase from hepatitis A virus (HAV) and implications for the polyprotein processing in HAV.
Virology, 265, 1999
|
|
