1OXX
 
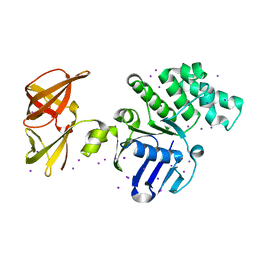 | | Crystal structure of GlcV, the ABC-ATPase of the glucose ABC transporter from Sulfolobus solfataricus | | Descriptor: | ABC transporter, ATP binding protein, IODIDE ION | | Authors: | Verdon, G, Albers, S.-V, van Oosterwijk, N, Dijkstra, B.W, Driessen, A.J.M, Thunnissen, A.M.W.H. | | Deposit date: | 2003-04-03 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Formation of the productive ATP-Mg2+-bound dimer of GlcV, an ABC-ATPase from Sulfolobus solfataricus
J.Mol.Biol., 334, 2003
|
|
2Y9W
 
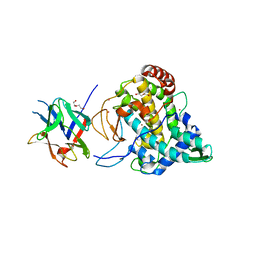 | | Crystal structure of PPO3, a tyrosinase from Agaricus bisporus, in deoxy-form that contains additional unknown lectin-like subunit | | Descriptor: | COPPER (II) ION, DI(HYDROXYETHYL)ETHER, HOLMIUM ATOM, ... | | Authors: | Ismaya, W.T, Rozeboom, H.J, Weijn, A, Mes, J.J, Fusetti, F, Wichers, H.J, Dijkstra, B.W. | | Deposit date: | 2011-02-17 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Agaricus Bisporus Mushroom Tyrosinase: Identity of the Tetramer Subunits and Interaction with Tropolone.
Biochemistry, 50, 2011
|
|
2YII
 
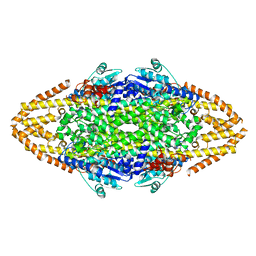 | | Manipulating the regioselectivity of phenylalanine aminomutase: new insights into the reaction mechanism of MIO-dependent enzymes from structure-guided directed evolution | | Descriptor: | BETA-MERCAPTOETHANOL, FORMIC ACID, GLYCEROL, ... | | Authors: | Wu, B, Szymanski, W, Wybenga, G.G, Heberling, M.M, Bartsch, S, Wildeman, S, Poelarends, G.J, Feringa, B.L, Dijkstra, B.W, Janssen, D.B. | | Deposit date: | 2011-05-13 | | Release date: | 2011-11-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Mechanism-Inspired Engineering of Phenylalanine Aminomutase for Enhanced Beta-Regioselective Asymmetric Amination of Cinnamates.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
2YFT
 
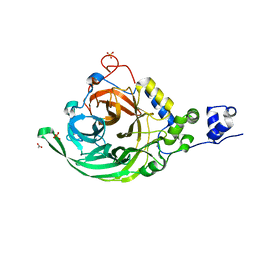 | | Crystal structure of inulosucrase from Lactobacillus johnsonii NCC533 in complex with 1-kestose | | Descriptor: | ACETATE ION, CALCIUM ION, LEVANSUCRASE, ... | | Authors: | Pijning, T, Anwar, M.A, Leemhuis, H, Kralj, S, Dijkhuizen, L, Dijkstra, B.W. | | Deposit date: | 2011-04-07 | | Release date: | 2011-08-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Inulosucrase from Lactobacillus: Insights Into the Substrate Specificity and Product Specificity of Gh68 Fructansucrases.
J.Mol.Biol., 412, 2011
|
|
2YFS
 
 | | Crystal structure of inulosucrase from Lactobacillus johnsonii NCC533 in complex with sucrose | | Descriptor: | CALCIUM ION, LEVANSUCRASE, SULFATE ION, ... | | Authors: | Pijning, T, Anwar, M.A, Leemhuis, H, Kralj, S, Dijkhuizen, L, Dijkstra, B.W. | | Deposit date: | 2011-04-07 | | Release date: | 2011-08-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Inulosucrase from Lactobacillus: Insights Into the Substrate Specificity and Product Specificity of Gh68 Fructansucrases.
J.Mol.Biol., 412, 2011
|
|
1S0Y
 
 | | The structure of trans-3-chloroacrylic acid dehalogenase, covalently inactivated by the mechanism-based inhibitor 3-bromopropiolate at 2.3 Angstrom resolution | | Descriptor: | MALONIC ACID, alpha-subunit of trans-3-chloroacrylic acid dehalogenase, beta-subunit of trans-3-chloroacrylic acid dehalogenase | | Authors: | de Jong, R.M, Brugman, W, Poelarends, G.J, Whitman, C.P, Dijkstra, B.W. | | Deposit date: | 2004-01-05 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The X-ray structure of trans-3-chloroacrylic acid dehalogenase reveals a novel hydration mechanism in the tautomerase superfamily
J.Biol.Chem., 279, 2004
|
|
1RY9
 
 | | Spa15, a Type III Secretion Chaperone from Shigella flexneri | | Descriptor: | CHLORIDE ION, Surface presentation of antigens protein spaK | | Authors: | van Eerde, A, Hamiaux, C, Perez, J, Parsot, C, Dijkstra, B.W. | | Deposit date: | 2003-12-20 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure of Spa15, a type III secretion chaperone from Shigella flexneri with broad specificity.
Embo Rep., 5, 2004
|
|
1D0K
 
 | | THE ESCHERICHIA COLI LYTIC TRANSGLYCOSYLASE SLT35 IN COMPLEX WITH TWO MURODIPEPTIDES (GLCNAC-MURNAC-L-ALA-D-GLU) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid, 35KD SOLUBLE LYTIC TRANSGLYCOSYLASE, ALANINE, ... | | Authors: | van Asselt, E.J, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 1999-09-12 | | Release date: | 2000-03-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystallographic studies of the interactions of Escherichia coli lytic transglycosylase Slt35 with peptidoglycan.
Biochemistry, 39, 2000
|
|
1CZF
 
 | | ENDO-POLYGALACTURONASE II FROM ASPERGILLUS NIGER | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, POLYGALACTURONASE II, ZINC ION | | Authors: | van Santen, Y, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 1999-09-02 | | Release date: | 1999-10-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | 1.68-A crystal structure of endopolygalacturonase II from Aspergillus niger and identification of active site residues by site-directed mutagenesis.
J.Biol.Chem., 274, 1999
|
|
1D0M
 
 | | THE ESCHERICHIA COLI LYTIC TRANSGLYCOSYLASE SLT35 IN COMPLEX WITH BULGECIN A AND (GLCNAC)2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, 35KD SOLUBLE LYTIC TRANSGLYCOSYLASE, 4-O-(4-O-SULFONYL-N-ACETYLGLUCOSAMININYL)-5-METHYLHYDROXY-L-PROLINE-TAURINE, ... | | Authors: | van Asselt, E.J, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 1999-09-12 | | Release date: | 2000-03-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystallographic studies of the interactions of Escherichia coli lytic transglycosylase Slt35 with peptidoglycan.
Biochemistry, 39, 2000
|
|
1A3A
 
 | | CRYSTAL STRUCTURE OF IIA MANNITOL FROM ESCHERICHIA COLI | | Descriptor: | MANNITOL-SPECIFIC EII | | Authors: | Van Montfort, R.L.M, Pijning, T, Kalk, K.H, Hangyi, I, Kouwijzer, M.L.C.E, Robillard, G.T, Dijkstra, B.W. | | Deposit date: | 1998-01-19 | | Release date: | 1998-08-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of the Escherichia coli phosphotransferase IIAmannitol reveals a novel fold with two conformations of the active site.
Structure, 6, 1998
|
|
1D0L
 
 | |
1A6J
 
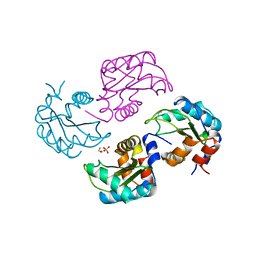 | | NITROGEN REGULATORY BACTERIAL PROTEIN IIA-NITROGEN | | Descriptor: | BETA-MERCAPTOETHANOL, NITROGEN REGULATORY IIA PROTEIN, SULFATE ION | | Authors: | Bordo, D, Van Montfort, R, Pijning, T, Kalk, K.H, Reizer, J, Saier, M.H, Dijkstra, B.W. | | Deposit date: | 1998-02-25 | | Release date: | 1998-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The three-dimensional structure of the nitrogen regulatory protein IIANtr from Escherichia coli.
J.Mol.Biol., 279, 1998
|
|
1KR0
 
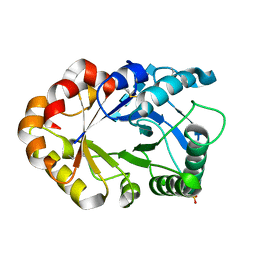 | | Hevamine Mutant D125A/Y183F in Complex with Tetra-NAG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hevamine A, SULFATE ION | | Authors: | Rozeboom, H.J, Dijkstra, B.W. | | Deposit date: | 2002-01-08 | | Release date: | 2002-01-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Expression and Characterization of Active Site Mutants of Hevamine, a
Chitinase from the Rubber Tree Hevea brasiliensis.
Eur.J.Biochem., 269, 2002
|
|
1KR1
 
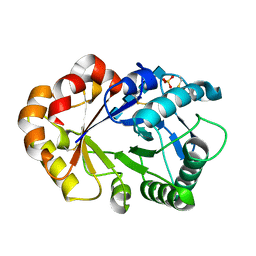 | | Hevamine Mutant D125A/E127A in Complex with Tetra-NAG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hevamine A, SULFATE ION | | Authors: | Rozeboom, H.J, Dijkstra, B.W. | | Deposit date: | 2002-01-08 | | Release date: | 2002-01-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Expression and Characterization of Active Site Mutants of Hevamine, a
Chitinase from the Rubber Tree Hevea brasiliensis.
Eur.J.Biochem., 269, 2002
|
|
1YOB
 
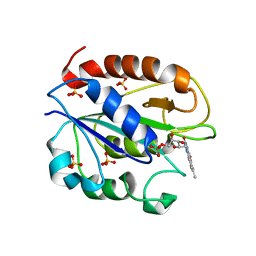 | | C69A Flavodoxin II from Azotobacter vinelandii | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin 2, SULFATE ION | | Authors: | Alagaratnam, S, van Pouderoyen, G, Pijning, T, Dijkstra, B.W, Cavazzini, D, Rossi, G.L, Canters, G.W. | | Deposit date: | 2005-01-27 | | Release date: | 2005-10-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A crystallographic study of Cys69Ala flavodoxin II from Azotobacter vinelandii: structural determinants of redox potential
Protein Sci., 14, 2005
|
|
1ZMT
 
 | | Structure of haloalcohol dehalogenase HheC of Agrobacterium radiobacter AD1 in complex with (R)-para-nitro styrene oxide, with a water molecule in the halide-binding site | | Descriptor: | (R)-PARA-NITROSTYRENE OXIDE, Haloalcohol dehalogenase HheC | | Authors: | de Jong, R.M, Tiesinga, J.J.W, Villa, A, Tang, L, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2005-05-10 | | Release date: | 2005-10-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for the Enantioselectivity of an Epoxide Ring Opening Reaction Catalyzed by Halo Alcohol Dehalogenase HheC.
J.Am.Chem.Soc., 127, 2005
|
|
1KQZ
 
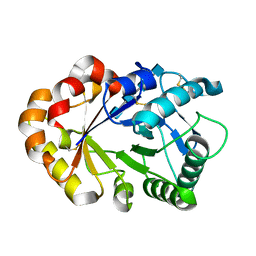 | | Hevamine Mutant D125A/E127A/Y183F in Complex with Tetra-NAG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hevamine A | | Authors: | Rozeboom, H.J, Dijkstra, B.W. | | Deposit date: | 2002-01-08 | | Release date: | 2002-01-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Expression and Characterization of Active Site Mutants of Hevamine, a
Chitinase from the Rubber Tree Hevea brasiliensis.
Eur.J.Biochem., 269, 2002
|
|
1KQY
 
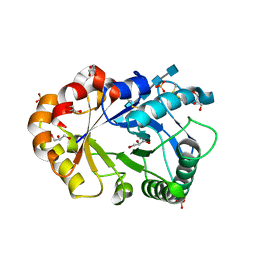 | | Hevamine Mutant D125A/E127A/Y183F in Complex with Penta-NAG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Hevamine A, ... | | Authors: | Rozeboom, H.J, Dijkstra, B.W. | | Deposit date: | 2002-01-08 | | Release date: | 2002-01-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Expression and characterization of active site mutants of hevamine, a chitinase from the rubber tree Hevea brasiliensis.
Eur.J.Biochem., 269, 2002
|
|
1ZMO
 
 | | Apo structure of haloalcohol dehalogenase HheA of Arthrobacter sp. AD2 | | Descriptor: | halohydrin dehalogenase | | Authors: | de Jong, R.M, Kalk, K.H, Tang, L, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2005-05-10 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The X-ray structure of the haloalcohol dehalogenase HheA from Arthrobacter sp. strain AD2: insight into enantioselectivity and halide binding in the haloalcohol dehalogenase family.
J.Bacteriol., 188, 2006
|
|
2AE0
 
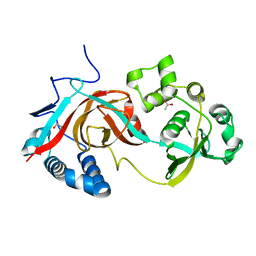 | | Crystal structure of MltA from Escherichia coli reveals a unique lytic transglycosylase fold | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Membrane-bound lytic murein transglycosylase A | | Authors: | Van Straaten, K.E, Dijkstra, B.W, Vollmer, W, Thunnissen, A.M.W.H. | | Deposit date: | 2005-07-21 | | Release date: | 2005-10-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of MltA from Escherichia coli Reveals a Unique Lytic Transglycosylase Fold
J.Mol.Biol., 352, 2005
|
|
2B4K
 
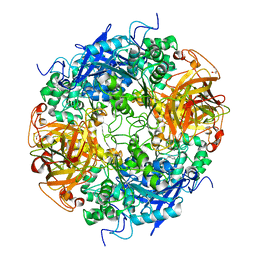 | | Acetobacter turbidans alpha-amino acid ester hydrolase complexed with phenylglycine | | Descriptor: | Alpha-amino acid ester hydrolase, D-PHENYLGLYCINE, GLYCEROL | | Authors: | Barends, T.R.M, Polderman-Tijmes, J.J, Jekel, P.A, Williams, C, Wybenga, G, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2005-09-26 | | Release date: | 2005-12-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Acetobacter turbidans alpha-amino acid ester hydrolase: how a single mutation improves an antibiotic-producing enzyme.
J.Biol.Chem., 281, 2006
|
|
1ZO8
 
 | | X-ray Structure of the haloalcohol dehalogenase HheC of Agrobacterium radiobacter AD1 in complex with (S)-para-nitrostyrene oxide, with a water molecule in the halide-binding site | | Descriptor: | (S)-PARA-NITROSTYRENE OXIDE, halohydrin dehalogenase | | Authors: | de Jong, R.M, Tiesinga, J.J.W, Tang, L, Villa, A, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2005-05-12 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Enantioselectivity of an Epoxide Ring Opening Reaction Catalyzed by Halo Alcohol Dehalogenase HheC.
J.Am.Chem.Soc., 127, 2005
|
|
1LLO
 
 | | HEVAMINE A (A PLANT ENDOCHITINASE/LYSOZYME) COMPLEXED WITH ALLOSAMIDIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-allopyranose, ALLOSAMIZOLINE, Hevamine-A | | Authors: | Terwisscha Van Scheltinga, A.C, Armand, S, Kalk, K.H, Isogai, A, Henrissat, B, Dijkstra, B.W. | | Deposit date: | 1995-11-08 | | Release date: | 1996-03-08 | | Last modified: | 2022-06-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Stereochemistry of chitin hydrolysis by a plant chitinase/lysozyme and X-ray structure of a complex with allosamidin: evidence for substrate assisted catalysis.
Biochemistry, 34, 1995
|
|
1ILD
 
 | | OUTER MEMBRANE PHOSPHOLIPASE A FROM ESCHERICHIA COLI N156A ACTIVE SITE MUTANT pH 4.6 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, OUTER MEMBRANE PHOSPHOLIPASE A, octyl beta-D-glucopyranoside | | Authors: | Snijder, H.J, Van Eerde, J.H, Kingma, R.L, Kalk, K.H, Dekker, N, Egmond, M.R, Dijkstra, B.W. | | Deposit date: | 2001-05-08 | | Release date: | 2001-10-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural investigations of the active-site mutant Asn156Ala of outer membrane phospholipase A: function of the Asn-His interaction in the catalytic triad.
Protein Sci., 10, 2001
|
|
