5K6J
 
 | | Human Phospodiesterase 4B in complex with pyridyloxy-benzoxaborole based inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 6-[[7,7-bis(oxidanyl)-8-oxa-7-boranuidabicyclo[4.3.0]nona-1(6),2,4-trien-3-yl]oxy]-5-chloranyl-2-(4-oxidanylidenepentoxy)pyridine-3-carbonitrile, MAGNESIUM ION, ... | | Authors: | Rock, F.L, Zhou, Y, Sullivan, D. | | Deposit date: | 2016-05-24 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | To be published
To Be Published
|
|
5JXD
 
 | | Crystal structure of murine Tnfaip8 C165S mutant | | Descriptor: | Tumor necrosis factor alpha-induced protein 8, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Park, J, Kim, M.S, Lee, D, Shin, D.H. | | Deposit date: | 2016-05-13 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.029 Å) | | Cite: | The Tnfaip8-PE complex is a novel upstream effector in the anti-autophagic action of insulin
Sci Rep, 7, 2017
|
|
5G2W
 
 | | S. enterica HisA mutant D10G, G102A | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, SULFATE ION | | Authors: | Guo, X, Soderholm, A, Newton, M, Nasvall, J, Andersson, D, Patrick, W, Selmer, M. | | Deposit date: | 2016-04-14 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural and functional innovations in the real-time evolution of new ( beta alpha )8 barrel enzymes.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5BWR
 
 | |
5KD7
 
 | | Crystal Structure of Murine MHC-I H-2Dd in complex with Murine Beta2-Microglobulin and a Variant of Peptide (PV9) of HIV gp120 MN Isolate (IGPGRAFYV) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Jiang, J, Natarajan, K, Margulies, D. | | Deposit date: | 2016-06-07 | | Release date: | 2017-10-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Effects of Cross-Presentation, Antigen Processing, and Peptide Binding in HIV Evasion of T Cell Immunity.
J. Immunol., 200, 2018
|
|
5K7C
 
 | | The native structure of native pistol ribozyme | | Descriptor: | DNA/RNA 11-MER, MAGNESIUM ION, RNA 47-MER | | Authors: | Ren, A, Patel, D. | | Deposit date: | 2016-05-26 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Pistol ribozyme adopts a pseudoknot fold facilitating site-specific in-line cleavage.
Nat.Chem.Biol., 12, 2016
|
|
5JSV
 
 | | tRNA guanine Transglycosylase (TGT) in co-crystallized complex with 6-amino-2-((((3aR,4R,6R,6aR)-6-methoxy-2,2-dimethyltetrahydrofuro[3,4-d][1,3]dioxol-4-yl)methyl)amino)-1H-imidazo[4,5-g]quinazolin-8(7H)-one | | Descriptor: | CHLORIDE ION, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Ehrmann, F.R, Nguyen, D, Heine, A, Klebe, G. | | Deposit date: | 2016-05-09 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Application of carbohydrates based lin-benzoguanines in a solvent-exposed subpocket of the tRNA-modifying Enzyme TGT
To be Published
|
|
5C7L
 
 | | Structure of human testis-specific glyceraldehyde-3-phosphate dehydrogenase apo form | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, testis-specific | | Authors: | Betts, L, Machius, M, Danshina, P, O'Brien, D. | | Deposit date: | 2015-06-24 | | Release date: | 2016-03-09 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.862 Å) | | Cite: | Structural analyses to identify selective inhibitors of glyceraldehyde 3-phosphate dehydrogenase-S, a sperm-specific glycolytic enzyme.
Mol. Hum. Reprod., 22, 2016
|
|
5K89
 
 | | Crystal Structure of Human Calcium-Bound S100A1 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Protein S100-A1 | | Authors: | Melville, Z, Aligholizadeh, E, McKnight, L.E, Weber, D, Pozharski, E, Weber, D.J. | | Deposit date: | 2016-05-27 | | Release date: | 2017-04-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | X-ray crystal structure of human calcium-bound S100A1.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5K5S
 
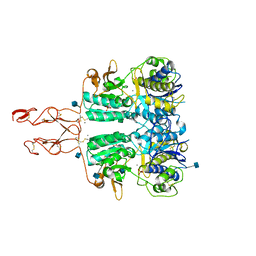 | | Crystal structure of the active form of human calcium-sensing receptor extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Extracellular calcium-sensing receptor, ... | | Authors: | Geng, Y, Mosyak, L, Kurinov, I, Zuo, H, Sturchler, E, Cheng, T.C, Subramanyam, P, Brown, A.P, Brennan, S.C, Mun, H.-C, Bush, M, Chen, Y, Nguyen, T, Cao, B, Chang, D, Quick, M, Conigrave, A, Colecraft, H.M, McDonald, P, Fan, Q.R. | | Deposit date: | 2016-05-23 | | Release date: | 2016-08-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural mechanism of ligand activation in human calcium-sensing receptor.
Elife, 5, 2016
|
|
5K7D
 
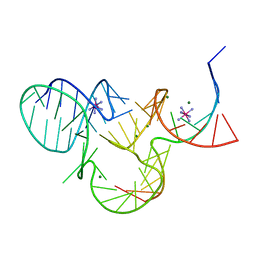 | | The structure of native pistol ribozyme, bound to Iridium | | Descriptor: | DNA/RNA 11-MER, IRIDIUM HEXAMMINE ION, MAGNESIUM ION, ... | | Authors: | Ren, A, Patel, D. | | Deposit date: | 2016-05-26 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Pistol ribozyme adopts a pseudoknot fold facilitating site-specific in-line cleavage.
Nat.Chem.Biol., 12, 2016
|
|
5KDD
 
 | | Apo-structure of humanised RadA-mutant humRadA22 | | Descriptor: | DNA repair and recombination protein RadA, SULFATE ION | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-06-08 | | Release date: | 2016-10-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5KDM
 
 | |
5KD4
 
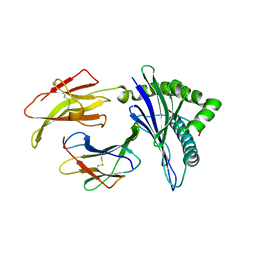 | | Crystal Structure of Murine MHC-I H-2Dd in complex with Murine Beta2-Microglobulin and a Variant of Peptide (PVI10) of HIV gp120 MN Isolate (IGPGRAFYVI) | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-D alpha chain, ... | | Authors: | Jiang, J, Natarajan, K, Margulies, D. | | Deposit date: | 2016-06-07 | | Release date: | 2017-10-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Effects of Cross-Presentation, Antigen Processing, and Peptide Binding in HIV Evasion of T Cell Immunity.
J. Immunol., 200, 2018
|
|
6AR4
 
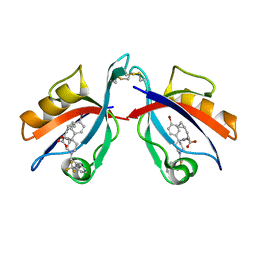 | | Crystal structure of PICK1 in complex with the small molecule inhibitor 1o | | Descriptor: | N-[4-(4-bromophenyl)-1-{[2-(trifluoromethyl)phenyl]methyl}piperidine-4-carbonyl]-3-cyclopropyl-L-alanine, PRKCA-binding protein | | Authors: | Marcotte, D. | | Deposit date: | 2017-08-21 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Potent PDZ-Domain PICK1 Inhibitors that Modulate Amyloid Beta-Mediated Synaptic Dysfunction.
Sci Rep, 8, 2018
|
|
5CLP
 
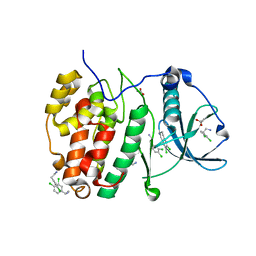 | | Crystal Structure of CK2alpha with 3,4-dichlorophenethylamine bound | | Descriptor: | 2-(3,4-dichlorophenyl)ethanamine, ACETATE ION, Casein kinase II subunit alpha | | Authors: | Brear, P, De Fusco, C, Georgiou, K.H, Spring, D, Hyvonen, M. | | Deposit date: | 2015-07-16 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.684 Å) | | Cite: | Specific inhibition of CK2 alpha from an anchor outside the active site.
Chem Sci, 7, 2016
|
|
8UKH
 
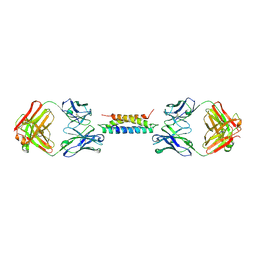 | |
5CSV
 
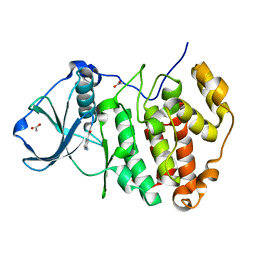 | | Crystal Structure of CK2alpha with Compound 6 bound | | Descriptor: | 3-AMINOBENZOIC ACID, ACETATE ION, Casein kinase II subunit alpha | | Authors: | Brear, P, De Fusco, C, Georgiou, K.H, Spring, D, Hyvonen, M. | | Deposit date: | 2015-07-23 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.375 Å) | | Cite: | Specific inhibition of CK2 alpha from an anchor outside the active site.
Chem Sci, 7, 2016
|
|
5CU4
 
 | | Crystal structure of CK2alpha bound to CAM4066 | | Descriptor: | ACETATE ION, Casein kinase II subunit alpha, N-[(2-chlorobiphenyl-4-yl)methyl]-beta-alanyl-N-(3-carboxyphenyl)-beta-alaninamide | | Authors: | Brear, P, De Fusco, C, Georgiou, K.H, Spring, D, Hyvonen, M. | | Deposit date: | 2015-07-24 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Specific inhibition of CK2 alpha from an anchor outside the active site.
Chem Sci, 7, 2016
|
|
5CVG
 
 | | Crystal Structure of CK2alpha with a novel closed conformation of the aD loop | | Descriptor: | ACETATE ION, Casein kinase II subunit alpha | | Authors: | Brear, P, De Fusco, C, Georgiou, K.H, Spring, D, Hyvonen, M. | | Deposit date: | 2015-07-26 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Specific inhibition of CK2 alpha from an anchor outside the active site.
Chem Sci, 7, 2016
|
|
6B7E
 
 | | Crystal structure of E.coli Phosphopantetheine Adenylyltransferase (PPAT/CoaD) in complex with (R)-4-(5-(difluoromethyl)-1H-imidazol-1-yl)-3,3-dimethylisochroman-1-one | | Descriptor: | (4R)-4-[5-(difluoromethyl)-1H-imidazol-1-yl]-3,3-dimethyl-3,4-dihydro-1H-2-benzopyran-1-one, DIMETHYL SULFOXIDE, Phosphopantetheine adenylyltransferase, ... | | Authors: | Proudfoot, A.W, Bussiere, D, Lingel, A. | | Deposit date: | 2017-10-03 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | High-Confidence Protein-Ligand Complex Modeling by NMR-Guided Docking Enables Early Hit Optimization.
J. Am. Chem. Soc., 139, 2017
|
|
5CU0
 
 | | Crystal structure of CK2alpha with 2-hydroxy-5-methylbenzoic acid and N-(3-(3-chloro-4-(phenyl)benzylamino)propyl)acetamide bound | | Descriptor: | 2-hydroxy-5-methylbenzoic acid, ACETATE ION, Casein kinase II subunit alpha, ... | | Authors: | Brear, P, De Fusco, C, Georgiou, K.H, Spring, D, Hyvonen, M. | | Deposit date: | 2015-07-24 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | A fragment-based approach leading to the discovery of a novel binding site and the selective CK2 inhibitor CAM4066.
Bioorg. Med. Chem., 25, 2017
|
|
5GT8
 
 | | Crystal Structure of apo-CASTOR1 | | Descriptor: | GATS-like protein 3 | | Authors: | Guo, L, Deng, D. | | Deposit date: | 2016-08-18 | | Release date: | 2017-08-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of arginine sensor CASTOR1 in arginine-bound and ligand free states
Biochem. Biophys. Res. Commun., 508, 2019
|
|
6B7F
 
 | | Crystal structure of E.coli Phosphopantetheine Adenylyltransferase (PPAT/CoaD) in complex with (R)-3,3-dimethyl-4-(5-vinyl-1H-imidazol-1-yl)isochroman-1-one | | Descriptor: | (4R)-4-(5-ethenyl-1H-imidazol-1-yl)-3,3-dimethyl-3,4-dihydro-1H-2-benzopyran-1-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Proudfoot, A.W, Bussiere, D, Lingel, A. | | Deposit date: | 2017-10-03 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.562 Å) | | Cite: | High-Confidence Protein-Ligand Complex Modeling by NMR-Guided Docking Enables Early Hit Optimization.
J. Am. Chem. Soc., 139, 2017
|
|
8X1F
 
 | |
