2NZI
 
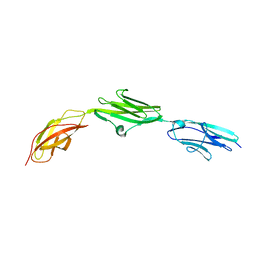 | |
2NY3
 
 | | HIV-1 gp120 Envelope Glycoprotein (K231C, T257S, E267C, S334A, S375W) Complexed with CD4 and Antibody 17b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY 17B, HEAVY CHAIN, ... | | Authors: | Zhou, T, Xu, L, Dey, B, Hessell, A.J, Van Ryk, D, Xiang, S.H, Yang, X, Zhang, M.Y, Zwick, M.B, Arthos, J, Burton, D.R, Dimitrov, D.S, Sodroski, J, Wyatt, R, Nabel, G.J, Kwong, P.D. | | Deposit date: | 2006-11-20 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural definition of a conserved neutralization epitope on HIV-1 gp120.
Nature, 445, 2007
|
|
2O1B
 
 | | Structure of aminotransferase from Staphylococcus aureus | | Descriptor: | Aminotransferase, class I, PYRIDOXAL-5'-PHOSPHATE | | Authors: | McGrath, T.E, Dharamsi, A, Thambipillai, D, Edwards, A.M, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2006-11-28 | | Release date: | 2006-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of aminotransferase from Staphylococcus aureus
To be Published
|
|
2O1Y
 
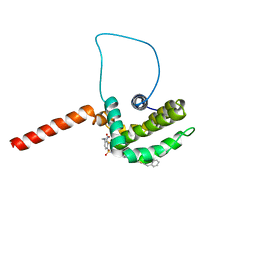 | | Solution structure of the anti-apoptotic protein Bcl-xL in complex with an acyl-sulfonamide-based ligand | | Descriptor: | 3-NITRO-N-{4-[2-(2-PHENYLETHYL)-1,3-BENZOTHIAZOL-5-YL]BENZOYL}-4-{[2-(PHENYLSULFANYL)ETHYL]AMINO}BENZENESULFONAMIDE, Apoptosis regulator Bcl-X | | Authors: | Bruncko, M, Oost, T.K, Belli, B.A, Ding, H, Joseph, M.K, Kunzer, A, Martineau, D, McClellan, W.J, Mitten, M, Ng, S.C, Nimmer, P.M, Oltersdorf, T, Park, C.M, Petros, A.M, Shoemaker, A.R, Song, X, Wang, X, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H, Elmore, S.W. | | Deposit date: | 2006-11-29 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Studies Leading to Potent, Dual Inhibitors of Bcl-2 and Bcl-xL.
J.Med.Chem., 50, 2007
|
|
1OZ9
 
 | | Crystal structure of AQ_1354, a hypothetical protein from Aquifex aeolicus | | Descriptor: | Hypothetical protein AQ_1354 | | Authors: | Oganesyan, V, Busso, D, Brandsen, J, Chen, S, Jancarik, J, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2003-04-08 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.894 Å) | | Cite: | Structure of the hypothetical protein AQ_1354 from Aquifex aeolicus.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
2O22
 
 | | Solution structure of the anti-apoptotic protein Bcl-2 in complex with an acyl-sulfonamide-based ligand | | Descriptor: | Apoptosis regulator Bcl-2, N-[(4-{[1,1-dimethyl-2-(phenylthio)ethyl]amino}-3-nitrophenyl)sulfonyl]-4-(4,4-dimethylpiperidin-1-yl)benzamide | | Authors: | Bruncko, M, Oost, T.K, Belli, B.A, Ding, H, Joseph, M.K, Kunzer, A, Martineau, D, McClellan, W.J, Mitten, M, Ng, S.C, Nimmer, P.M, Oltersdorf, T, Park, C.M, Petros, A.M, Shoemaker, A.R, Song, X, Wang, X, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H, Elmore, S.W. | | Deposit date: | 2006-11-29 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Studies Leading to Potent, Dual Inhibitors of Bcl-2 and Bcl-xL.
J.Med.Chem., 50, 2007
|
|
2O2M
 
 | | Solution structure of the anti-apoptotic protein Bcl-xL in complex with an acyl-sulfonamide-based ligand | | Descriptor: | 4-(4-BENZYL-4-METHOXYPIPERIDIN-1-YL)-N-[(4-{[1,1-DIMETHYL-2-(PHENYLTHIO)ETHYL]AMINO}-3-NITROPHENYL)SULFONYL]BENZAMIDE, Apoptosis regulator Bcl-X | | Authors: | Bruncko, M, Oost, T.K, Belli, B.A, Ding, H, Joseph, M.K, Kunzer, A, Martineau, D, McClellan, W.J, Mitten, M, Ng, S.C, Nimmer, P.M, Oltersdorf, T, Park, C.M, Petros, A.M, Shoemaker, A.R, Song, X, Wang, X, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H, Elmore, S.W. | | Deposit date: | 2006-11-30 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Studies Leading to Potent, Dual Inhibitors of Bcl-2 and Bcl-xL.
J.Med.Chem., 50, 2007
|
|
2O6G
 
 | |
3HOH
 
 | | RIBONUCLEASE T1 (THR93GLN MUTANT) COMPLEXED WITH 2'GMP | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, PROTEIN (RIBONUCLEASE T1) | | Authors: | Langhorst, U, Loris, R, Denisov, V.P, Doumen, J, Roose, P, Maes, D, Halle, B, Steyaert, J. | | Deposit date: | 1998-09-11 | | Release date: | 1998-09-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Dissection of the structural and functional role of a conserved hydration site in RNase T1.
Protein Sci., 8, 1999
|
|
3I1U
 
 | | Carboxypeptidase A Inhibited by a Thiirane Mechanism-Based inactivator | | Descriptor: | (2S,3R)-2-benzyl-3-sulfanylbutanoic acid, Carboxypeptidase A1 (Pancreatic), GLYCEROL, ... | | Authors: | Fernandez, D. | | Deposit date: | 2009-06-28 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.391 Å) | | Cite: | The X-ray structure of carboxypeptidase A inhibited by a thiirane mechanism-based inhibitor
Chem.Biol.Drug Des., 75, 2010
|
|
1P46
 
 | | T4 lysozyme core repacking mutant M106I/TA | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME, ... | | Authors: | Mooers, B.H, Datta, D, Baase, W.A, Zollars, E.S, Mayo, S.L, Matthews, B.W. | | Deposit date: | 2003-04-21 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Repacking the Core of T4 lysozyme by automated design
J.Mol.Biol., 332, 2003
|
|
3HOY
 
 | | Complete RNA polymerase II elongation complex VI | | Descriptor: | 5'-D(*CP*CP*AP*AP*GP*CP*TP*CP*AP*AP*G*TP*AP*CP*TP*TP*AP*CP*GP*CP*CP*(BRU)P*GP*GP*TP*CP*AP*TP*TP*AP*CP*TP*AP*GP*TP*AP*CP*TP*GP*CP*C)-3', 5'-D(*CP*CP*GP*GP*CP*AP*GP*TP*AP*CP*TP*AP*GP*TP*AP*AP*AP*CP*TP*AP*GP*TP*AP*TP*T*GP*AP*AP*AP*GP*TP*AP*CP*TP*TP*GP*AP*GP*CP*TP*T)-3', 5'-R(*UP*AP*UP*AP*UP*GP*CP*A*UP*AP*AP*AP*GP*AP*CP*CP*AP*GP*GP*A)-3', ... | | Authors: | Sydow, J.F, Brueckner, F, Cheung, A.C.M, Damsma, G.E, Dengl, S, Lehmann, E, Vassylyev, D, Cramer, P. | | Deposit date: | 2009-06-03 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis of transcription: mismatch-specific fidelity mechanisms and paused RNA polymerase II with frayed RNA.
Mol.Cell, 34, 2009
|
|
1OZ3
 
 | | Crystal Structure of 3-MBT repeats of lethal (3) malignant Brain Tumor (Native-I) at 1.85 angstrom | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lethal(3)malignant brain tumor-like protein, SULFATE ION | | Authors: | Wang, W.K, Tereshko, V, Boccuni, P, MacGrogan, D, Nimer, S.D, Patel, D.J. | | Deposit date: | 2003-04-07 | | Release date: | 2003-08-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Malignant brain tumor repeats: a three-leaved propeller architecture with ligand/peptide binding pockets.
Structure, 11, 2003
|
|
3I32
 
 | |
4OAL
 
 | | Crystal structure of maize cytokinin oxidase/dehydrogenase 4 (ZmCKO4) in complex with phenylurea inhibitor CPPU in alternative spacegroup | | Descriptor: | 1-(2-chloropyridin-4-yl)-3-phenylurea, Cytokinin dehydrogenase 4, DIMETHYL SULFOXIDE, ... | | Authors: | Kopecny, D, Morera, S, Vigouroux, A, Koncitikova, R. | | Deposit date: | 2014-01-05 | | Release date: | 2015-04-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Kinetic and structural investigation of the cytokinin oxidase/dehydrogenase active site.
Febs J., 283, 2016
|
|
1P7S
 
 | | T4 LYSOZYME CORE REPACKING MUTANT V103I/TA | | Descriptor: | LYSOZYME | | Authors: | Mooers, B.H, Datta, D, Baase, W.A, Zollars, E.S, Mayo, S.L, Matthews, B.W. | | Deposit date: | 2003-05-05 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Repacking the Core of T4 Lysozyme by Automated Design
J.Mol.Biol., 332, 2003
|
|
2NRH
 
 | | Crystal structure of conserved putative Baf family transcriptional activator from Campylobacter jejuni | | Descriptor: | SULFATE ION, Transcriptional activator, putative, ... | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Lau, C, Wasserman, S, Smith, D, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-02 | | Release date: | 2006-11-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of conserved putative Baf family transcriptional activator from Campylobacter jejuni
To be Published
|
|
3HV6
 
 | | Human p38 MAP Kinase in Complex with RL39 | | Descriptor: | 1-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]-3-[4-(2-morpholin-4-ylethoxy)phenyl]urea, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Getlik, M, Rauh, D. | | Deposit date: | 2009-06-15 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Displacement assay for the detection of stabilizers of inactive kinase conformations.
J.Med.Chem., 53, 2010
|
|
1P94
 
 | | NMR Structure of ParG symmetric dimer | | Descriptor: | plasmid partition protein ParG | | Authors: | Golovanov, A.P, Barilla, D, Golovanova, M, Hayes, F, Lian, L.Y. | | Deposit date: | 2003-05-09 | | Release date: | 2004-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | ParG, a protein required for active partition of bacterial plasmids, has a dimeric ribbon-helix-helix structure.
Mol.Microbiol., 50, 2003
|
|
3HVZ
 
 | | Crystal Structure of the TGS domain of the CLOLEP_03100 protein from Clostridium leptum, Northeast Structural Genomics Consortium Target QlR13A | | Descriptor: | Uncharacterized protein | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-16 | | Release date: | 2009-06-23 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Northeast Structural Genomics Consortium Target QlR13A
To be Published
|
|
2NT1
 
 | | Structure of acid-beta-glucosidase at neutral pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glucosylceramidase, PHOSPHATE ION | | Authors: | Lieberman, R.L, Petsko, G.A, Ringe, D. | | Deposit date: | 2006-11-06 | | Release date: | 2006-12-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of acid beta-glucosidase with pharmacological chaperone provides insight into Gaucher disease.
Nat.Chem.Biol., 3, 2007
|
|
1ONB
 
 | |
1OPO
 
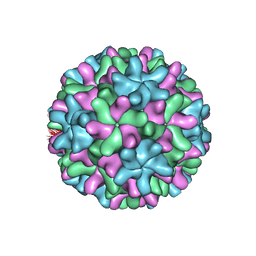 | | THE STRUCTURE OF CARNATION MOTTLE VIRUS | | Descriptor: | CALCIUM ION, Coat protein, SULFATE ION | | Authors: | Morgunova, E, Dauter, Z, Fry, E, Stuart, D, Stel'mashchuk, V, Mikhailov, A.M, Wilson, K.S, Vainshtein, B.K. | | Deposit date: | 2003-03-06 | | Release date: | 2003-04-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The atomic structure of Carnation Mottle Virus capsid protein
Febs Lett., 338, 1994
|
|
2NR4
 
 | | Crystal structure of FMN-bound protein MM1853 from Methanosarcina mazei, Pfam DUF447 | | Descriptor: | Conserved hypothetical protein, FLAVIN MONONUCLEOTIDE | | Authors: | Bonanno, J.B, Gilmore, J, Bain, K.T, Lau, C, Wasserman, S, Smith, D, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-01 | | Release date: | 2006-11-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of conserved FMN bound hypothetical protein from Methanosarcina mazei
To be Published
|
|
1P6S
 
 | | Solution Structure of the Pleckstrin Homology Domain of Human Protein Kinase B beta (Pkb/Akt) | | Descriptor: | RAC-beta serine/threonine protein kinase | | Authors: | Auguin, D, Barthe, P, Auge-Senegas, M.T, Stern, M.H, Noguchi, M, Roumestand, C. | | Deposit date: | 2003-04-30 | | Release date: | 2004-05-18 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of the pleckstrin homology domain of the human
protein kinase B (PKB/Akt). Interaction with inositol phosphates.
J.BIOMOL.NMR, 28, 2004
|
|
