7MVW
 
 | | Crystal structure of Chaetomium thermophilum Nup188 NTD (residues 1-1134) | | Descriptor: | GLYCEROL, Nucleoporin NUP188 | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-05-15 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
7MVT
 
 | | Crystal structure of the Chaetomium thermophilum Nup192-Nic96 complex (Nup192 residues 185-1756; Nic96 residues 187-301) | | Descriptor: | Nucleoporin NIC96, Nucleoporin NUP192 | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-05-15 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
7MVV
 
 | | Single particle cryo-EM structure of the Chaetomium thermophilum Nup192-Nic96-Nup53-Nup145N complex (Nup192 residues 1-1756; Nic96 residues 240-301; Nup53 31-67; Nup145N 616-683) | | Descriptor: | Nucleoporin NIC96, Nucleoporin NUP145N, Nucleoporin NUP192, ... | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-05-15 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
7YXA
 
 | | XFEL crystal structure of the human sphingosine 1 phosphate receptor 5 in complex with ONO-5430608 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[6-(2-naphthalen-1-ylethoxy)-2,3,4,5-tetrahydro-1H-3-benzazepin-3-ium-3-yl]butanoic acid, ... | | Authors: | Lyapina, E, Marin, E, Gusach, A, Orekhov, P, Gerasimov, A, Luginina, A, Vakhrameev, D, Ergasheva, M, Kovaleva, M, Khusainov, G, Khorn, P, Shevtsov, M, Kovalev, K, Okhrimenko, I, Bukhdruker, S, Popov, P, Hu, H, Weierstall, U, Liu, W, Cho, Y, Gushchin, I, Rogachev, A, Bourenkov, G, Park, S, Park, G, Huyn, H.J, Park, J, Gordeliy, V, Borshchevskiy, V, Mishin, A, Cherezov, V. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for receptor selectivity and inverse agonism in S1P 5 receptors.
Nat Commun, 13, 2022
|
|
7MVX
 
 | | Crystal structure of the Chaetomium thermophilum Nup188-Nic96 complex (Nup188 residues 1-1858; Nic96 residues 240-301) | | Descriptor: | Nucleoporin NIC96, Nucleoporin NUP188 | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-05-15 | | Release date: | 2022-06-15 | | Last modified: | 2022-06-22 | | Method: | X-RAY DIFFRACTION (4.35 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
7YX5
 
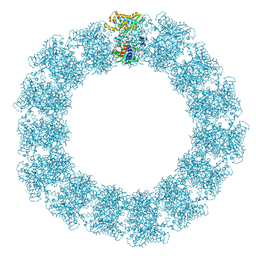 | | Structure of the Mimivirus genomic fibre in its relaxed 5-start helix form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative glucose-methanol-choline oxidoreductase protein | | Authors: | Villalta, A, Schmitt, A, Estrozi, L.F, Quemin, E.R.J, Alempic, J.M, Lartigue, A, Prazak, V, Belmudes, L, Vasishtan, D, Colmant, A.M.G, Honore, F.A, Coute, Y, Grunewald, K, Abergel, C. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The giant mimivirus 1.2 Mb genome is elegantly organized into a 30 nm diameter helical protein shield.
Elife, 11, 2022
|
|
7MVU
 
 | | Single particle cryo-EM structure of the Chaetomium thermophilum Nup192-Nic96 complex (Nup192 residues 1-1756; Nic96 residues 240-301) | | Descriptor: | Nucleoporin NIC96, Nucleoporin NUP192 | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-05-15 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
7MVY
 
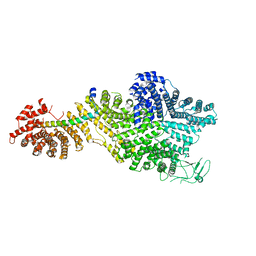 | | Single particle cryo-EM structure of the Chaetomium thermophilum Nup188-Nic96 complex (Nup188 residues 1-1858; Nic96 residues 240-301) | | Descriptor: | Nucleoporin NIC96, Nucleoporin NUP188 | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-05-15 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
7YX4
 
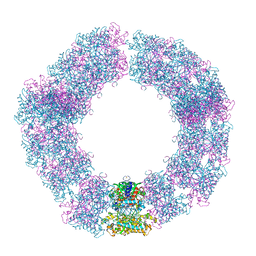 | | Structure of the Mimivirus genomic fibre in its compact 5-start helix form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative glucose-methanol-choline oxidoreductase protein | | Authors: | Villalta, A, Schmitt, A, Estrozi, L.F, Quemin, E.R.J, Alempic, J.M, Lartigue, A, Prazak, V, Belmudes, L, Vasishtan, D, Colmant, A.M.G, Honore, F.A, Coute, Y, Grunewald, K, Abergel, C. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The giant mimivirus 1.2 Mb genome is elegantly organized into a 30 nm diameter helical protein shield.
Elife, 11, 2022
|
|
7MW1
 
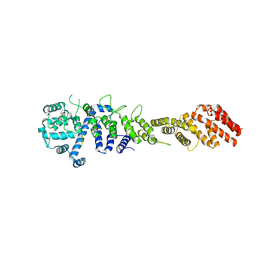 | | Crystal structure of the Homo sapiens NUP93-NUP53 complex (NUP93 residues 174-819; NUP53 residues 84-150) | | Descriptor: | Nuclear pore complex protein Nup93, Nucleoporin Nup35 | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-05-15 | | Release date: | 2022-06-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
7MW0
 
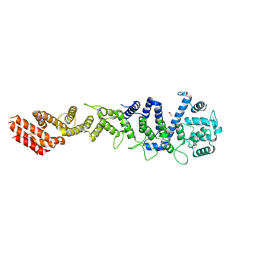 | | Crystal structure of Homo sapiens NUP93 solenoid (residues 174-819) | | Descriptor: | 1,2-ETHANEDIOL, Nuclear pore complex protein Nup93 | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-05-15 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
7YX3
 
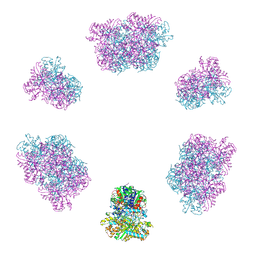 | | Structure of the Mimivirus genomic fibre in its compact 6-start helix form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative GMC-type oxidoreductase | | Authors: | Villalta, A, Schmitt, A, Estrozi, L.F, Quemin, E.R.J, Alempic, J.M, Lartigue, A, Prazak, V, Belmudes, L, Vasishtan, D, Colmant, A.M.G, Honore, F.A, Coute, Y, Grunewald, K, Abergel, C. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The giant mimivirus 1.2 Mb genome is elegantly organized into a 30-nm diameter helical protein shield.
Elife, 11, 2022
|
|
7ZC8
 
 | |
7M5T
 
 | |
4BOH
 
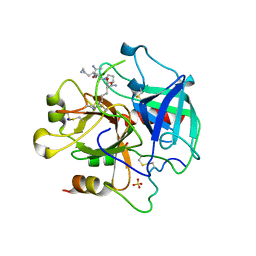 | | Madanins (MEROPS I53) are cleaved by thrombin and factor Xa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, SULFATE ION, ... | | Authors: | Figueiredo, A.C, deSanctis, D, Pereira, P.J.B. | | Deposit date: | 2013-05-20 | | Release date: | 2013-09-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | The Tick-Derived Anticoagulant Madanin is Processed by Thrombin and Factor Xa.
Plos One, 8, 2013
|
|
7MP7
 
 | | Rules for designing protein fold switches and their implications for the folding code | | Descriptor: | Sb3 | | Authors: | He, Y, Chen, Y, Ruan, B, Choi, J, Chen, Y, Motabar, D, Solomon, T, Simmerman, R, Kauffman, T, Gallagher, T, Bryan, P, Orban, J. | | Deposit date: | 2021-05-04 | | Release date: | 2022-05-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design and characterization of a protein fold switching network.
Nat Commun, 14, 2023
|
|
7MN1
 
 | | Rules for designing protein fold switches and their implications for the folding code | | Descriptor: | Sa1 | | Authors: | He, Y, Chen, Y, Ruan, B, Choi, J, Chen, Y, Motabar, D, Solomon, T, Simmerman, R, Kauffman, T, Gallagher, T, Bryan, P, Orban, J. | | Deposit date: | 2021-04-30 | | Release date: | 2022-05-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design and characterization of a protein fold switching network.
Nat Commun, 14, 2023
|
|
7M1W
 
 | |
7MQ4
 
 | | Rules for designing protein fold switches and their implications for the folding code | | Descriptor: | Sb1 | | Authors: | He, Y, Chen, Y, Ruan, B, Choi, J, Chen, Y, Motabar, D, Solomon, T, Simmerman, R, Kauffman, T, Gallagher, T, Bryan, P, Orban, J. | | Deposit date: | 2021-05-05 | | Release date: | 2022-05-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design and characterization of a protein fold switching network.
Nat Commun, 14, 2023
|
|
4EYT
 
 | | Crystal structure of the C-terminal domain of Tetrahymena telomerase protein p65 | | Descriptor: | SULFATE ION, Telomerase associated protein p65 | | Authors: | Singh, M, Wang, Z, Koo, B.-K, Patel, A, Cascio, D, Collins, K, Feigon, J. | | Deposit date: | 2012-05-01 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Telomerase RNA Recognition and RNP Assembly by the Holoenzyme La Family Protein p65.
Mol.Cell, 47, 2012
|
|
7NAY
 
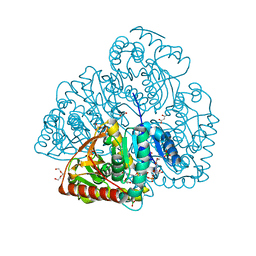 | | Crystal structure of lactate dehydrogenase from Selenomonas ruminantium with NADH. | | Descriptor: | 1,2-ETHANEDIOL, BORIC ACID, L-lactate dehydrogenase, ... | | Authors: | Bertrand, Q, Robin, A, Girard, E, Madern, D. | | Deposit date: | 2021-01-25 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Biochemical, structural and dynamical characterizations of the lactate dehydrogenase from Selenomonas ruminantium provide information about an intermediate evolutionary step prior to complete allosteric regulation acquisition in the super family of lactate and malate dehydrogenases.
J.Struct.Biol., 215, 2023
|
|
3RK3
 
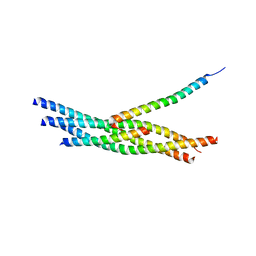 | | Truncated SNARE complex with complexin | | Descriptor: | Complexin-1, SNAP25, Syntaxin 1a, ... | | Authors: | Kuemmel, D, Reinisch, K.M. | | Deposit date: | 2011-04-17 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Complexin cross-links prefusion SNAREs into a zigzag array.
Nat.Struct.Mol.Biol., 18, 2011
|
|
7NE1
 
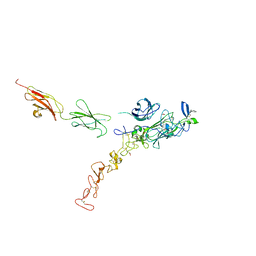 | | Structure of the complex between Netrin-1 and its receptor Neogenin | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Robinson, R.A, Griffiths, S.C, van de Haar, L.L, Malinauskas, T, van Battum, E.Y, Zelina, P, Schwab, R.A, Karia, D, Malinauskaite, L, Brignani, S, van den Munkhof, M, Dudukcu, O, De Ruiter, A.A, Van den Heuvel, D.M.A, Bishop, B, Elegheert, J, Aricescu, A.R, Pasterkamp, R.J, Siebold, C. | | Deposit date: | 2021-02-02 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Simultaneous binding of Guidance Cues NET1 and RGM blocks extracellular NEO1 signaling.
Cell, 184, 2021
|
|
7NDG
 
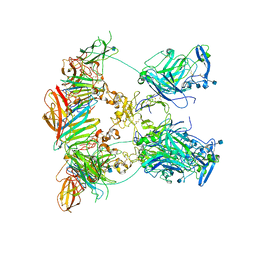 | | Cryo-EM structure of the ternary complex between Netrin-1, Neogenin and Repulsive Guidance Molecule B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neogenin, ... | | Authors: | Robinson, R.A, Griffiths, S.C, van de Haar, L.L, Malinauskas, T, van Battum, E.Y, Zelina, P, Schwab, R.A, Karia, D, Malinauskaite, L, Brignani, S, van den Munkhof, M, Dudukcu, O, De Ruiter, A.A, Van den Heuvel, D.M.A, Bishop, B, Elegheert, J, Aricescu, A.R, Pasterkamp, R.J, Siebold, C. | | Deposit date: | 2021-02-01 | | Release date: | 2021-03-31 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (5.98 Å) | | Cite: | Simultaneous binding of Guidance Cues NET1 and RGM blocks extracellular NEO1 signaling.
Cell, 184, 2021
|
|
7NE0
 
 | | Structure of the ternary complex between Netrin-1, Repulsive-Guidance Molecule-B (RGMB) and Neogenin | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Robinson, R.A, Griffiths, S.C, van de Haar, L.L, Malinauskas, T, van Battum, E.Y, Zelina, P, Schwab, R.A, Karia, D, Malinauskaite, L, Brignani, S, van den Munkhof, M, Dudukcu, O, De Ruiter, A.A, Van den Heuvel, D.M.A, Bishop, B, Elegheert, J, Aricescu, A.R, Pasterkamp, R.J, Siebold, C. | | Deposit date: | 2021-02-02 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Simultaneous binding of Guidance Cues NET1 and RGM blocks extracellular NEO1 signaling.
Cell, 184, 2021
|
|
