3FI1
 
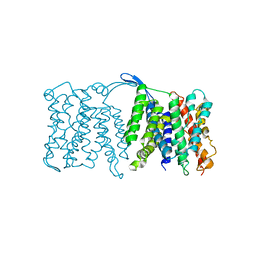 | | NhaA dimer model | | Descriptor: | Na(+)/H(+) antiporter nhaA | | Authors: | Appel, M, Hizlan, D, Vinothkumar, K.R, Ziegler, C, Kuehlbrandt, W. | | Deposit date: | 2008-12-10 | | Release date: | 2009-01-13 | | Last modified: | 2024-02-21 | | Method: | ELECTRON CRYSTALLOGRAPHY (7 Å) | | Cite: | Conformations of NhaA, the Na/H exchanger from Escherichia coli, in the pH-activated and ion-translocating states
J.Mol.Biol., 386, 2009
|
|
6VA9
 
 | |
6UZN
 
 | |
6UZS
 
 | |
3F9R
 
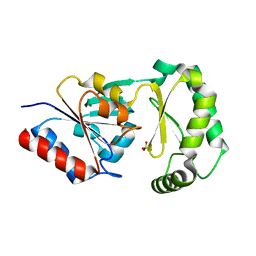 | | Crystal Structure of Trypanosoma Brucei phosphomannosemutase, TB.10.700.370 | | Descriptor: | MAGNESIUM ION, Phosphomannomutase, SULFATE ION | | Authors: | Wernimont, A.K, Lam, A, Ali, A, Lin, Y.H, Guther, L, Shamshad, A, MacKenzie, F, Bandini, G, Kozieradzki, I, Cossar, D, Zhao, Y, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Ferguson, M.A.J, Hui, R, Qiu, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-14 | | Release date: | 2009-03-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Trypanosoma Brucei phosphomannosemutase, TB.10.700.370
To be Published
|
|
6TDA
 
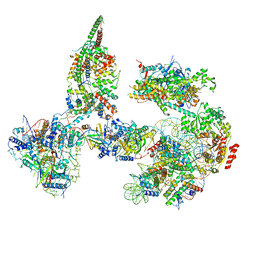 | | Structure of SWI/SNF chromatin remodeler RSC bound to a nucleosome | | Descriptor: | Actin-like protein ARP9, Actin-related protein 7, Chromatin structure-remodeling complex protein RSC58, ... | | Authors: | Wagner, F.R, Dienemann, C, Wang, H, Stuetzer, A, Tegunov, D, Urlaub, H, Cramer, P. | | Deposit date: | 2019-11-08 | | Release date: | 2020-03-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Structure of SWI/SNF chromatin remodeller RSC bound to a nucleosome.
Nature, 579, 2020
|
|
6TE9
 
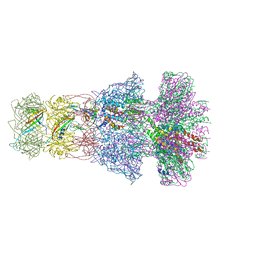 | | Neck of native GTA particle computed with C6 symmetry | | Descriptor: | Adaptor protein Rcc01688, Phage major tail protein, TP901-1 family, ... | | Authors: | Bardy, P, Fuzik, T, Hrebik, D, Pantucek, R, Beatty, J.T, Plevka, P. | | Deposit date: | 2019-11-11 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structure and mechanism of DNA delivery of a gene transfer agent.
Nat Commun, 11, 2020
|
|
6TE2
 
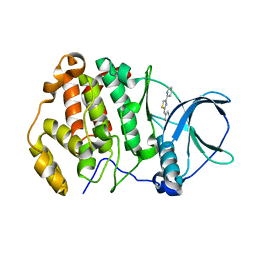 | | Crystal structure of human protein kinase CK2alpha' (CSNK2A2 gene product) in complex with the 2-aminothiazole-type inhibitor 17 | | Descriptor: | 3-[(4-pyridin-2-yl-1,3-thiazol-2-yl)amino]benzoic acid, Casein kinase II subunit alpha' | | Authors: | Niefind, K, Lindenblatt, D, Jose, J, Applegate, V.M, Nickelsen, A. | | Deposit date: | 2019-11-11 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (0.922 Å) | | Cite: | Structural and Mechanistic Basis of the Inhibitory Potency of Selected 2-Aminothiazole Compounds on Protein Kinase CK2.
J.Med.Chem., 63, 2020
|
|
6TE8
 
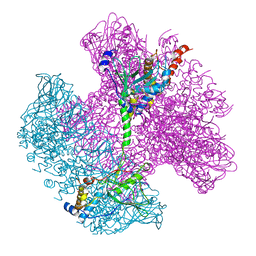 | | Neck of native GTA particle computed with C12 symmetry | | Descriptor: | Adaptor protein Rcc01688, Phage portal protein, HK97 family | | Authors: | Bardy, P, Fuzik, T, Hrebik, D, Pantucek, R, Beatty, J.T, Plevka, P. | | Deposit date: | 2019-11-11 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structure and mechanism of DNA delivery of a gene transfer agent.
Nat Commun, 11, 2020
|
|
6TEB
 
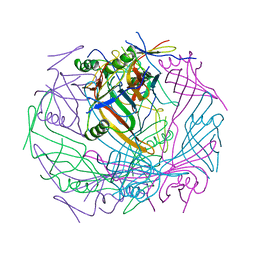 | | Tail-baseplate interface of native GTA particle computed with C6 symmetry | | Descriptor: | Distal tail protein Rcc01695, Tail tube protein Rcc01691 | | Authors: | Bardy, P, Fuzik, T, Hrebik, D, Pantucek, R, Beatty, J.T, Plevka, P. | | Deposit date: | 2019-11-11 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Structure and mechanism of DNA delivery of a gene transfer agent.
Nat Commun, 11, 2020
|
|
6TFZ
 
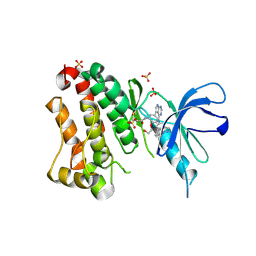 | | Crystal Structure of EGFR T790M/V948R in Complex with Covalent Pyrrolopyrimidine 19 | | Descriptor: | 1,2-ETHANEDIOL, Epidermal growth factor receptor, SULFATE ION, ... | | Authors: | Niggenaber, J, Mueller, M.P, Rauh, D. | | Deposit date: | 2019-11-14 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Targeting Her2-insYVMA with Covalent Inhibitors-A Focused Compound Screening and Structure-Based Design Approach.
J.Med.Chem., 63, 2020
|
|
6TG7
 
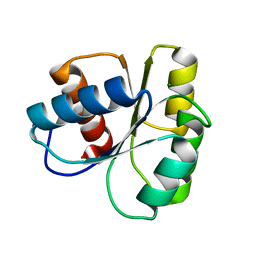 | |
6TFA
 
 | |
6TL0
 
 | | Solution structure and 1H, 13C and 15N chemical shift assignments for the complex of VPS29 with VARP 687-747 | | Descriptor: | Ankyrin repeat domain-containing protein 27, Vacuolar protein sorting-associated protein 29, ZINC ION | | Authors: | Owen, D.J, Neuhaus, D, Yang, J.-C, Crawley-Snowdon, H. | | Deposit date: | 2019-11-29 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mechanism and evolution of the Zn-fingernail required for interaction of VARP with VPS29.
Nat Commun, 11, 2020
|
|
6TLJ
 
 | |
6TOA
 
 | | Neck of empty GTA particle computed with C6 symmetry | | Descriptor: | Adaptor protein Rcc01688, Portal protein Rcc01684, Stopper protein Rcc01689, ... | | Authors: | Bardy, P, Fuzik, T, Hrebik, D, Pantucek, R, Beatty, J.T, Plevka, P. | | Deposit date: | 2019-12-11 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structure and mechanism of DNA delivery of a gene transfer agent.
Nat Commun, 11, 2020
|
|
3EMN
 
 | | The Crystal Structure of Mouse VDAC1 at 2.3 A resolution | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Voltage-dependent anion-selective channel protein 1 | | Authors: | Ujwal, R, Cascio, D, Colletier, J.-P, Faham, S, Zhang, J, Toro, L, Ping, P, Abramson, J. | | Deposit date: | 2008-09-24 | | Release date: | 2008-12-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of mouse VDAC1 at 2.3 A resolution reveals mechanistic insights into metabolite gating
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
6TTW
 
 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 4 (ASI_M3M_047) | | Descriptor: | (2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)-~{N}-piperidin-4-yl-oxolane-2-carboxamide, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-12-30 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Small-Molecule Inhibitors of METTL3, the Major Human Epitranscriptomic Writer.
Chemmedchem, 15, 2020
|
|
6TSQ
 
 | | Marasmius oreades agglutinin (MOA) activated by manganese (II) | | Descriptor: | 1,2-ETHANEDIOL, Agglutinin, CALCIUM ION, ... | | Authors: | Cordara, G, Manna, D, Krengel, U. | | Deposit date: | 2019-12-21 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of MOA in complex with a peptide fragment: A protease caught in flagranti .
Curr Res Struct Biol, 2, 2020
|
|
3ELZ
 
 | | Crystal structure of Zebrafish Ileal Bile Acid-Bindin Protein complexed with cholic acid (crystal form A). | | Descriptor: | CHOLIC ACID, ileal Bile Acid-Binding Protein | | Authors: | Capaldi, S, Saccomani, G, Fessas, D, Signorelli, M, Perduca, M, Monaco, H.L. | | Deposit date: | 2008-09-23 | | Release date: | 2009-01-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The X-Ray structure of zebrafish (Danio rerio) ileal bile acid-binding protein reveals the presence of binding sites on the surface of the protein molecule.
J.Mol.Biol., 385, 2009
|
|
6TTY
 
 | | Structure of ClpP from Staphylococcus aureus (apo, closed state) | | Descriptor: | ATP-dependent Clp protease proteolytic subunit | | Authors: | Malik, I.T, Pereira, R, Vielberg, M.-T, Mayer, C, Straetener, J, Thomy, D, Famulla, K, Castro, H.C, Sass, P, Groll, M, Broetz-Oesterheldt, H. | | Deposit date: | 2019-12-30 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Functional Characterisation of ClpP Mutations Conferring Resistance to Acyldepsipeptide Antibiotics in Firmicutes.
Chembiochem, 21, 2020
|
|
6TLA
 
 | | CRYSTAL STRUCTURE OF LECTIN-LIKE OX-LDL RECEPTOR 1 (C 1 2 1) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Oxidized low-density lipoprotein receptor 1 | | Authors: | Nar, H, Fiegen, D, Schnapp, G. | | Deposit date: | 2019-12-02 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | A small-molecule inhibitor of lectin-like oxidized LDL receptor-1 acts by stabilizing an inactive receptor tetramer state
Commun Chem, 2020
|
|
3ELX
 
 | | Crystal structure of apo Zebrafish Ileal Bile Acid-Binding Protein | | Descriptor: | 1,2-ETHANEDIOL, Ileal bile acid-binding protein | | Authors: | Capaldi, S, Saccomani, G, Fessas, D, Signorelli, M, Perduca, M, Monaco, H.L. | | Deposit date: | 2008-09-23 | | Release date: | 2009-01-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The X-Ray structure of zebrafish (Danio rerio) ileal bile acid-binding protein reveals the presence of binding sites on the surface of the protein molecule.
J.Mol.Biol., 385, 2009
|
|
6SRW
 
 | | Kemp Eliminase HG3.17 mutant Q50F, E47N, N300D Complexed with Transition State Analog 6-Nitrobenzotriazole | | Descriptor: | 6-NITROBENZOTRIAZOLE, GLYCEROL, Kemp Eliminase HG3.17 Q50F | | Authors: | Bloch, J.S, Pinkas, D.M, Hilvert, D. | | Deposit date: | 2019-09-06 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Contribution of Oxyanion Stabilization to Kemp Eliminase Efficiencyproficiency
Acs Catalysis, 2020
|
|
3E80
 
 | |
