4ABW
 
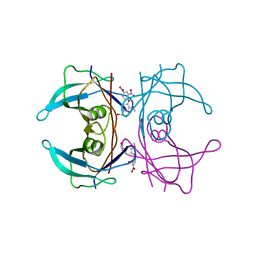 | | Crystal Structure of Transthyretin in Complex With Ligand C-6 | | Descriptor: | TRANSTHYRETIN, {4-[4-(hydroxymethyl)-2-methoxyphenoxy]benzene-1,3-diyl}bis[hydroxy(oxo)ammonium] | | Authors: | Tomar, D, Khan, T, Singh, R.R, Mishra, S, Gupta, S, Surolia, A, Salunke, D.M. | | Deposit date: | 2011-12-11 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic Study of Novel Transthyretin Ligands Exhibiting Negative-Cooperativity between Two Thyroxine Binding Sites.
Plos One, 7, 2012
|
|
6RPC
 
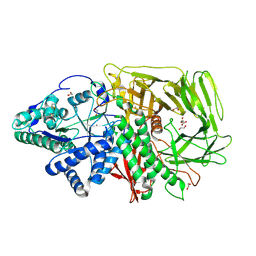 | | GOLGI ALPHA-MANNOSIDASE II | | Descriptor: | 1,2-ETHANEDIOL, Alpha-mannosidase 2, SUCCINIC ACID, ... | | Authors: | Armstrong, Z, Lahav, D, Johnson, R, Kuo, C.L, Beenakker, T.J.M, de Boer, C, Wong, C.S, Debets, M, van der Stelt, M, Codee, J.D.C, Aerts, J.M.F.G, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2019-05-14 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Manno- epi -cyclophellitols Enable Activity-Based Protein Profiling of Human alpha-Mannosidases and Discovery of New Golgi Mannosidase II Inhibitors.
J.Am.Chem.Soc., 142, 2020
|
|
4AM5
 
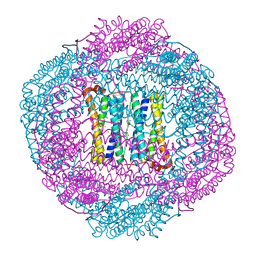 | | Bacterioferritin from Blastochloris viridis | | Descriptor: | BACTERIOFERRITIN, FE (III) ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wahlgren, W.Y, Omran, H, von Stetten, D, Royant, A, van der Post, S, Katona, G. | | Deposit date: | 2012-03-07 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural Characterization of Bacterioferritin from Blastochloris Viridis.
Plos One, 7, 2012
|
|
4AMG
 
 | | Crystal structure of the glycosyltransferase SnogD from Streptomyces nogalater | | Descriptor: | SNOGD | | Authors: | Claesson, M, Siitonen, V, Dobritzsch, D, Metsa-Ketela, M, Schneider, G. | | Deposit date: | 2012-03-09 | | Release date: | 2012-09-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal Structure of the Glycosyltransferase Snogd from the Biosynthetic Pathway of the Nogalamycin in Streptomyces Nogalater.
FEBS J., 279, 2012
|
|
6R3F
 
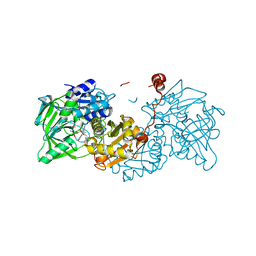 | |
4AOW
 
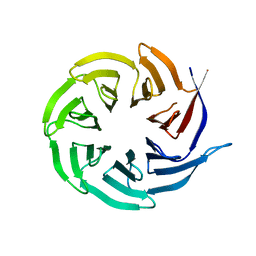 | | Crystal structure of the human Rack1 protein at a resolution of 2.45 angstrom | | Descriptor: | GLYCEROL, GUANINE NUCLEOTIDE-BINDING PROTEIN SUBUNIT BETA-2-LIKE 1 | | Authors: | Ruiz Carrillo, D, Chandrasekaran, R, Nilsson, M, Cornvick, T, Liew, C.W, Tan, S.M, Lescar, J. | | Deposit date: | 2012-03-30 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of Human Rack1 Protein at a Resolution of 2.45 A.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4ACX
 
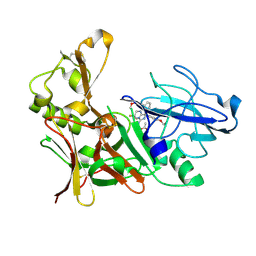 | | Aminoimidazoles as BACE-1 Inhibitors. X-RAY CRYSTAL STRUCTURE OF BETA SECRETASE COMPLEXED WITH COMPOUND 23 | | Descriptor: | (8R)-8-[4-(DIFLUOROMETHOXY)PHENYL]-3,3-DIFLUORO-8-[3-(3-METHOXYPROP-1-YN-1-YL)PHENYL]-2,3,4,8-TETRAHYDROIMIDAZO[1,5-A]PYRIMIDIN-6-AMINE, ACETATE ION, BETA-SECRETASE 1 | | Authors: | Swahn, B, Holenz, J, Kihlstrom, J, Kolmodin, K, Lindstrom, J, Plobeck, N, Rotticci, D, Sehgelmeble, F, Sundstrom, M, von Berg, S, Falting, J, Georgievska, B, Gustavsson, S, Neelissen, J, Ek, M, Olsson, L.L, Berg, S. | | Deposit date: | 2011-12-20 | | Release date: | 2012-02-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Aminoimidazoles as BACE-1 inhibitors: the challenge to achieve in vivo brain efficacy.
Bioorg. Med. Chem. Lett., 22, 2012
|
|
6RAF
 
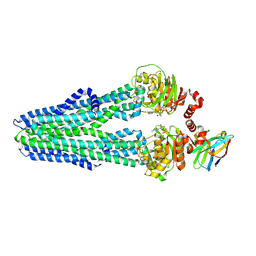 | | Heterodimeric ABC exporter TmrAB in inward-facing narrow conformation under turnover conditions | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Anti-vesicular stomatitis virus N VHH, ... | | Authors: | Thomas, C, Januliene, D, Mehdipour, A.R, Hofmann, S, Hummer, G, Moeller, A, Tampe, R. | | Deposit date: | 2019-04-06 | | Release date: | 2019-07-31 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Conformation space of a heterodimeric ABC exporter under turnover conditions.
Nature, 571, 2019
|
|
4ACU
 
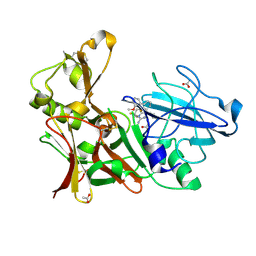 | | Aminoimidazoles as BACE-1 Inhibitors. X-RAY CRYSTAL STRUCTURE OF BETA SECRETASE COMPLEXED WITH COMPOUND 14 | | Descriptor: | (8S)-3,3-DIFLUORO-8-(2'-FLUORO-3'-METHOXYBIPHENYL-3-YL)-8-PYRIDIN-4-YL-2,3,4,8-TETRAHYDROIMIDAZO[1,5-A]PYRIMIDIN-6-AMINE, ACETATE ION, BETA-SECRETASE 1 | | Authors: | Swahn, B, Holenz, J, Kihlstrom, J, Kolmodin, K, Lindstrom, J, Plobeck, N, Rotticci, D, Sehgelmeble, F, Sundstrom, M, von Berg, S, Falting, J, Georgievska, B, Gustavsson, S, Neelissen, J, Ek, M, Olsson, L.L, Berg, S. | | Deposit date: | 2011-12-19 | | Release date: | 2012-02-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Aminoimidazoles as BACE-1 inhibitors: the challenge to achieve in vivo brain efficacy.
Bioorg. Med. Chem. Lett., 22, 2012
|
|
4A0R
 
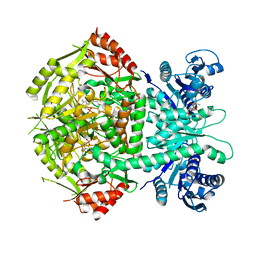 | | Structure of bifunctional DAPA aminotransferase-DTB synthetase from Arabidopsis thaliana bound to dethiobiotin (DTB). | | Descriptor: | 6-(5-METHYL-2-OXO-IMIDAZOLIDIN-4-YL)-HEXANOIC ACID, ADENOSYLMETHIONINE-8-AMINO-7-OXONONANOATE AMINOTRANSFERASE, L(+)-TARTARIC ACID, ... | | Authors: | Cobessi, D, Dumas, R, Pautre, V, Meinguet, C, Ferrer, J.L, Alban, C. | | Deposit date: | 2011-09-12 | | Release date: | 2012-06-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Biochemical and Structural Characterization of the Arabidopsis Bifunctional Enzyme Dethiobiotin Synthetase-Diaminopelargonic Acid Aminotransferase: Evidence for Substrate Channeling in Biotin Synthesis.
Plant Cell, 24, 2012
|
|
6RAL
 
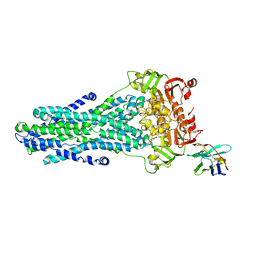 | | Heterodimeric ABC exporter TmrAB under turnover conditions in asymmetric unlocked return conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Thomas, C, Januliene, D, Mehdipour, A.R, Hofmann, S, Hummer, G, Moeller, A, Tampe, R. | | Deposit date: | 2019-04-06 | | Release date: | 2019-07-31 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Conformation space of a heterodimeric ABC exporter under turnover conditions.
Nature, 571, 2019
|
|
4A95
 
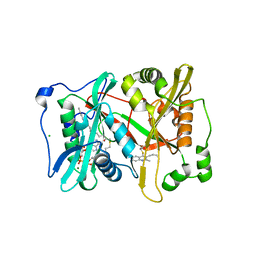 | | Plasmodium vivax N-myristoyltransferase with quinoline inhibitor | | Descriptor: | 2-oxopentadecyl-CoA, 3-(3-BUTYL-6-METHOXY-2-METHYL-QUINOLIN-4-YL)SULFANYLPROPANENITRILE, CHLORIDE ION, ... | | Authors: | Goncalves, V, Brannigan, J.A, Whalley, D, Ansell, K.H, Saxty, B, Holder, A.A, Wilkinson, A.J, Tate, E.W, Leatherbarrow, R.J. | | Deposit date: | 2011-11-24 | | Release date: | 2012-06-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of Plasmodium Vivax N-Myristoyltransferase Inhibitors: Screening, Synthesis, and Structural Characterization of Their Binding Mode.
J.Med.Chem., 55, 2012
|
|
6R43
 
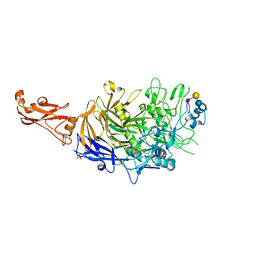 | | Structure of P110 from Mycoplasma Genitalium complexed with 6'-SL | | Descriptor: | Mgp-operon protein 3, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, PHOSPHATE ION, ... | | Authors: | Aparicio, D, Fita, I. | | Deposit date: | 2019-03-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Mycoplasma genitalium adhesin P110 binds sialic-acid human receptors.
Nat Commun, 9, 2018
|
|
6R6W
 
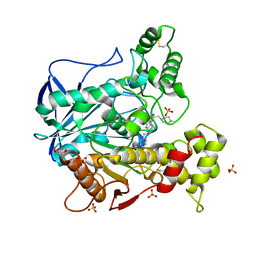 | | Structure of recombinant human butyrylcholinesterase in complex with a fluorescent NBD-based probe | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Cholinesterase, ... | | Authors: | Brazzolotto, X, Nachon, F, Knez, D, Gobec, S. | | Deposit date: | 2019-03-28 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.474 Å) | | Cite: | Development of potent reversible selective inhibitors of butyrylcholinesterase as fluorescent probes.
J Enzyme Inhib Med Chem, 35, 2020
|
|
4AMB
 
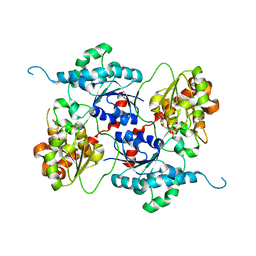 | | Crystal structure of the glycosyltransferase SnogD from Streptomyces nogalater | | Descriptor: | DEOXYURIDINE-5'-DIPHOSPHATE, SNOGD | | Authors: | Claesson, M, Siitonen, V, Dobritzsch, D, Metsa-Ketela, M, Schneider, G. | | Deposit date: | 2012-03-08 | | Release date: | 2012-09-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Crystal Structure of the Glycosyltransferase Snogd from the Biosynthetic Pathway of the Nogalamycin in Streptomyces Nogalater.
FEBS J., 279, 2012
|
|
4AOF
 
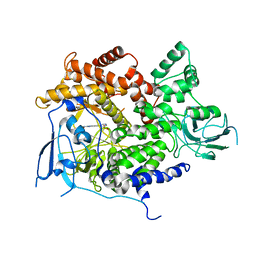 | | Selective small molecule inhibitor discovered by chemoproteomic assay platform reveals regulation of Th17 cell differentiation by PI3Kgamma | | Descriptor: | N-[6-(5-methylsulfonylpyridin-3-yl)-[1,2,4]triazolo[1,5-a]pyridin-2-yl]ethanamide, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM | | Authors: | Bergamini, G, Bell, K, Shimamura, S, Werner, T, Cansfield, A, Muller, K, Perrin, J, Rau, C, Ellard, K, Hopf, C, Doce, C, Leggate, D, Mangano, R, Mathieson, T, OMahony, A, Plavec, I, Rharbaoui, F, Reinhard, F, Savitski, M.M, Ramsden, N, Hirsch, E, Drewes, G, Rausch, O, Bantscheff, M, Neubauer, G. | | Deposit date: | 2012-03-26 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A Selective Inhibitor Reveals Pi3Kgamma Dependence of T(H)17 Cell Differentiation.
Nat.Chem.Biol., 8, 2012
|
|
4AA6
 
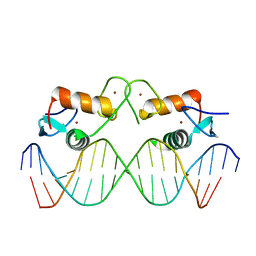 | | The oestrogen receptor recognizes an imperfectly palindromic response element through an alternative side-chain conformation | | Descriptor: | 5'-D(*CP*TP*AP*AP*GP*TP*CP*AP*CP*AP*GP*TP*GP*AP *CP*CP*TP*G)-3', 5'-D(*TP*CP*AP*GP*GP*TP*CP*AP*CP*TP*GP*TP*GP*AP *CP*TP*TP*A)-3', ESTROGEN RECEPTOR, ... | | Authors: | Schwabe, J.W, Chapman, L, Rhodes, D. | | Deposit date: | 2011-11-30 | | Release date: | 2011-12-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The oestrogen receptor recognizes an imperfectly palindromic response element through an alternative side-chain conformation.
Structure, 3, 1995
|
|
4AB3
 
 | | ATP-triggered molecular mechanics of the chaperonin GroEL | | Descriptor: | 60 KDA CHAPERONIN, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Clare, D.K, Vasishtan, D, Stagg, S, Quispe, J, Farr, G.W, Topf, M, Horwich, A.L, Saibil, H.R. | | Deposit date: | 2011-12-06 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | ATP-Triggered Conformational Changes Delineate Substrate-Binding and -Folding Mechanics of the Groel Chaperonin.
Cell(Cambridge,Mass.), 149, 2012
|
|
4ADX
 
 | | The Cryo-EM Structure of the Archaeal 50S Ribosomal Subunit in Complex with Initiation Factor 6 | | Descriptor: | 23S Ribosomal RNA EXPANSION SEGMENTS, 23S ribosomal RNA, 5S Ribosomal RNA, ... | | Authors: | Greber, B.J, Boehringer, D, Godinic-Mikulcic, V, Crnkovic, A, Ibba, M, Weygand-Durasevic, I, Ban, N. | | Deposit date: | 2012-01-04 | | Release date: | 2012-02-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Cryo-Em Structure of the Archaeal 50S Ribosomal Subunit in Complex with Initiation Factor 6 and Implications for Ribosome Evolution
J.Mol.Biol., 418, 2012
|
|
4AJF
 
 | | Identification and structural characterization of PDE10 fragment inhibitors | | Descriptor: | CAMP AND CAMP-INHIBITED CGMP 3', 5'-CYCLIC PHOSPHODIESTERASE 10A, MAGNESIUM ION, ... | | Authors: | Johansson, P, Albert, J.S, Spadola, L, Akerud, T, Back, E, Hillertz, P, Horsefeld, R, Scott, C, Spear, N, Tian, G, Tigerstrom, A, Aharony, D, Geschwindner, S. | | Deposit date: | 2012-02-16 | | Release date: | 2013-03-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification and Structural Characterization of Pde10 Fragment Inhibitors
To be Published
|
|
4AKA
 
 | | IPSE alpha-1, an IgE-binding crystallin | | Descriptor: | IL-4-INDUCING PROTEIN | | Authors: | Meyer, N.H, Mayerhofer, H, Tripsianes, K, Barths, D, Blindow, S, Bade, S, Madl, T, Frey, A, Haas, H, Mueller-Dieckmann, J, Sattler, M, Scharmm, G. | | Deposit date: | 2012-02-22 | | Release date: | 2013-03-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A Crystallin Fold in the Interleukin-4-Inducing Principle of Schistosoma Mansoni Eggs (Ipse/Alpha-1) Mediates Ige Binding for Antigen-Independent Basophil Activation
J.Biol.Chem., 290, 2015
|
|
4A4E
 
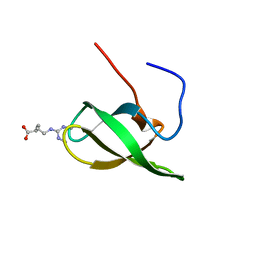 | | Solution structure of SMN Tudor domain in complex with symmetrically dimethylated arginine | | Descriptor: | N3, N4-DIMETHYLARGININE, SURVIVAL MOTOR NEURON PROTEIN | | Authors: | Tripsianes, K, Madl, T, Machyna, M, Fessas, D, Englbrecht, C, Fischer, U, Neugebauer, K.M, Sattler, M. | | Deposit date: | 2011-10-12 | | Release date: | 2011-11-30 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Dimethyl-Arginine Recognition by the Tudor Domains of Human Smn and Spf30 Proteins
Nat.Struct.Mol.Biol., 18, 2011
|
|
4A5V
 
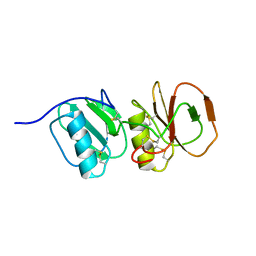 | | Solution structure ensemble of the two N-terminal apple domains (residues 58-231) of Toxoplasma gondii microneme protein 4 | | Descriptor: | MICRONEMAL PROTEIN 4 | | Authors: | Marchant, J, Cowper, B, Liu, Y, Lai, L, Pinzan, C, Marq, J.B, Friedrich, N, Sawmynaden, K, Chai, W, Childs, R.A, Saouros, S, Simpson, P, Barreira, M.C.R, Feizi, T, Soldati-Favre, D, Matthews, S. | | Deposit date: | 2011-10-28 | | Release date: | 2012-04-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Galactose Recognition by the Apicomplexan Parasite Toxoplasma Gondii.
J.Biol.Chem., 287, 2012
|
|
6QZ0
 
 | | The cryo-EM structure of the head of the genome empited bacteriophage phi29 | | Descriptor: | Capsid fiber protein, Major capsid protein | | Authors: | Xu, J, Wang, D, Gui, M, Xiang, Y. | | Deposit date: | 2019-03-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural assembly of the tailed bacteriophage φ29.
Nat Commun, 10, 2019
|
|
4A4H
 
 | | Solution structure of SPF30 Tudor domain in complex with asymmetrically dimethylated arginine | | Descriptor: | NG,NG-DIMETHYL-L-ARGININE, SURVIVAL OF MOTOR NEURON-RELATED-SPLICING FACTOR 30 | | Authors: | Tripsianes, K, Madl, T, Machyna, M, Fessas, D, Englbrecht, C, Fischer, U, Neugebauer, K.M, Sattler, M. | | Deposit date: | 2011-10-12 | | Release date: | 2011-11-30 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Dimethyl-Arginine Recognition by the Tudor Domains of Human Smn and Spf30 Proteins
Nat.Struct.Mol.Biol., 18, 2011
|
|
