1Y9I
 
 | | Crystal structure of low temperature requirement C protein from Listeria monocytogenes | | Descriptor: | CALCIUM ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-12-15 | | Release date: | 2004-12-28 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of phosphatidylglycerophosphatase (PGPase), a putative membrane-bound lipid phosphatase, reveals a novel binuclear metal binding site and two "proton wires".
Proteins, 64, 2006
|
|
1HOV
 
 | | SOLUTION STRUCTURE OF A CATALYTIC DOMAIN OF MMP-2 COMPLEXED WITH SC-74020 | | Descriptor: | CALCIUM ION, MATRIX METALLOPROTEINASE-2, N-{4-[(1-HYDROXYCARBAMOYL-2-METHYL-PROPYL)-(2-MORPHOLIN-4-YL-ETHYL)-SULFAMOYL]-4-PENTYL-BENZAMIDE, ... | | Authors: | Feng, Y, Likos, J.J, Zhu, L, Woodward, H, Munie, G, McDonald, J.J, Stevens, A.M, Howard, C.P, De Crescenzo, G.A, Welsch, D, Shieh, H.-S, Stallings, W.C. | | Deposit date: | 2000-12-11 | | Release date: | 2001-12-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of the catalytic domain of matrix metalloproteinase-2 complexed with a hydroxamic acid inhibitor
Biochim.Biophys.Acta, 1598, 2002
|
|
1HRF
 
 | | SOLUTION STRUCTURE OF THE EPIDERMAL GROWTH FACTOR-LIKE DOMAIN OF HEREGULIN-ALPHA, A LIGAND FOR P180ERB4 | | Descriptor: | HEREGULIN ALPHA | | Authors: | Nagata, K, Kohda, D, Hatanaka, H, Ichikawa, S, Inagaki, F. | | Deposit date: | 1994-07-21 | | Release date: | 1994-10-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the epidermal growth factor-like domain of heregulin-alpha, a ligand for p180erbB-4.
EMBO J., 13, 1994
|
|
1Y7H
 
 | | Structural and biochemical studies identify tobacco SABP2 as a methylsalicylate esterase and further implicate it in plant innate immunity, Northeast Structural Genomics Target AR2241 | | Descriptor: | THIOCYANATE ION, salicylic acid-binding protein 2 | | Authors: | Forouhar, F, Yang, Y, Kumar, D, Chen, Y, Fridman, E, Park, S.W, Chiang, Y, Acton, T.B, Montelione, G.T, Pichersky, E, Klessig, D.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-12-08 | | Release date: | 2004-12-28 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural and biochemical studies identify tobacco SABP2 as a methyl salicylate esterase and implicate it in plant innate immunity
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1HUJ
 
 | |
2A92
 
 | | Crystal structure of lactate dehydrogenase from Plasmodium vivax: complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-lactate dehydrogenase | | Authors: | Chaikuad, A, Fairweather, V, Conners, R, Joseph-Horne, T, Turgut-Balik, D, Brady, R.L. | | Deposit date: | 2005-07-11 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of Lactate Dehydrogenase from Plasmodium vivax: Complexes with NADH and APADH.
Biochemistry, 44, 2005
|
|
2AAI
 
 | | Crystallographic refinement of ricin to 2.5 Angstroms | | Descriptor: | RICIN (A CHAIN), RICIN (B CHAIN), alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rutenber, E, Katzin, B.J, Montfort, W, Villafranca, J.E, Ernst, S.R, Collins, E.J, Mlsna, D, Monzingo, A.F, Ready, M.P, Robertus, J.D. | | Deposit date: | 1993-09-07 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic refinement of ricin to 2.5 A.
Proteins, 10, 1991
|
|
1I36
 
 | | Structure of Conserved Protein MTH1747 of Unknown Function Reveals Structural Similarity with 3-Hydroxyacid Dehydrogenases | | Descriptor: | CONSERVED HYPOTHETICAL PROTEIN MTH1747, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Korolev, S.V, Dementieva, I.S, Christendat, D, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-02-13 | | Release date: | 2002-05-15 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | STRUCTURAL SIMILARITIES OF MTH1747 HYPOTHETICAL PROTEIN FROM METHANOBACTERIUM THERMOAUTOTROPHICUM WITH 3-HYDROXYACID DEHYDROGENASES
to be published
|
|
1IBH
 
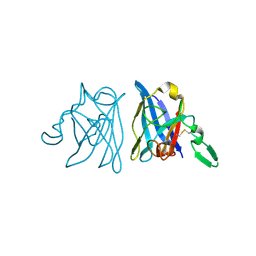 | | X-RAY 3D STRUCTURE OF P.LEIOGNATHI CU,ZN SOD MUTANT M41I | | Descriptor: | COPPER (II) ION, CU,ZN SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Stroppolo, M.E, Pesce, A, D'Orazio, M, O'Neill, P, Bordo, D, Rosano, C, Milani, M, Battistoni, A, Bolognesi, M, Desideri, A. | | Deposit date: | 2001-03-28 | | Release date: | 2001-05-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Single mutations at the subunit interface modulate copper reactivity in Photobacterium leiognathi Cu,Zn superoxide dismutase.
J.Mol.Biol., 308, 2001
|
|
2ABB
 
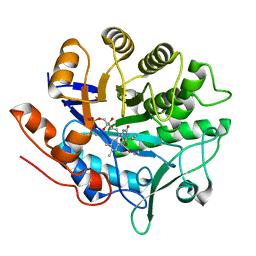 | | Structure of PETN reductase Y186F in complex with cyanide | | Descriptor: | FLAVIN MONONUCLEOTIDE, ISOPROPYL ALCOHOL, THIOCYANATE ION, ... | | Authors: | Khan, H, Barna, T, Bruce, N.C, Munro, A.W, Leys, D, Scrutton, N.S. | | Deposit date: | 2005-07-15 | | Release date: | 2005-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Proton transfer in the oxidative half-reaction of pentaerythritol tetranitrate reductase
Febs J., 272, 2005
|
|
1W5X
 
 | | HIV-1 protease in complex with fluoro substituted diol-based C2- symmetric inhibitor | | Descriptor: | (2R,3R,4R,5R)-2,5-BIS[(2,3-DIFLUOROBENZYL)OXY]-3,4-DIHYDROXY-N,N'-BIS[(1S,2R)-2-HYDROXY-2,3-DIHYDRO-1H-INDEN-1-YL]HEXAN EDIAMIDE, POL POLYPROTEIN | | Authors: | Lindberg, J, Pyring, D, Loewgren, S, Rosenquist, A, Zuccarello, G, Kvarnstroem, I, Zhang, H, Vrang, L, Claesson, B, Hallberg, A, Samuelsson, B, Unge, T. | | Deposit date: | 2004-08-10 | | Release date: | 2004-12-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Symmetric Fluoro-Substituted Diol-Based HIV Protease Inhibitors. Ortho-Fluorinated and Meta-Fluorinated P1/P1'-Benzyloxy Side Groups Significantly Improve the Antiviral Activity and Preserve Binding Efficacy
Eur.J.Biochem., 271, 2004
|
|
1W6U
 
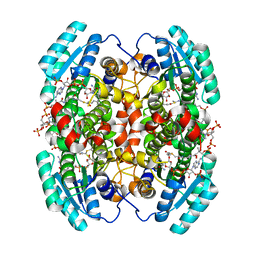 | | Structure of human DECR ternary complex | | Descriptor: | 2,4-DIENOYL-COA REDUCTASE, MITOCHONDRIAL PRECURSOR, HEXANOYL-COENZYME A, ... | | Authors: | Alphey, M.S, Byres, E, Li, D, Hunter, W.N. | | Deposit date: | 2004-08-24 | | Release date: | 2004-10-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and Reactivity of Human Mitochondrial 2,4-Dienoyl-Coa Reductase: Enzyme-Ligand Interactions in a Distinctive Short-Chain Reductase Active Site
J.Biol.Chem., 280, 2005
|
|
1IBB
 
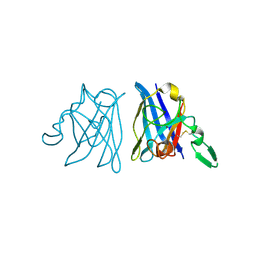 | | X-RAY 3D STRUCTURE OF P.LEIOGNATHI CU,ZN SOD MUTANT W83F | | Descriptor: | COPPER (II) ION, CU,ZN SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Stroppolo, M.E, Pesce, A, D'Orazio, M, O'Neill, P, Bordo, D, Rosano, C, Milani, M, Battistoni, A, Bolognesi, M, Desideri, A. | | Deposit date: | 2001-03-28 | | Release date: | 2001-05-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Single mutations at the subunit interface modulate copper reactivity in Photobacterium leiognathi Cu,Zn superoxide dismutase.
J.Mol.Biol., 308, 2001
|
|
2AE8
 
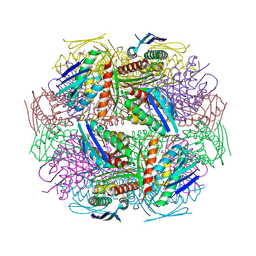 | | Crystal Structure of Imidazoleglycerol-phosphate Dehydratase from Staphylococcus aureus subsp. aureus N315 | | Descriptor: | Imidazoleglycerol-phosphate dehydratase, MAGNESIUM ION | | Authors: | Kim, Y, Quartey, P, Holzle, D, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-07-21 | | Release date: | 2005-09-06 | | Last modified: | 2015-05-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Structure of Imidazoleglycerol-phosphate Dehydratase from Staphylococcus aureus subsp. aureus N315
To be Published
|
|
1IBF
 
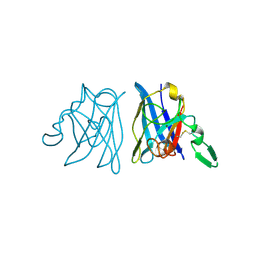 | | X-RAY 3D STRUCTURE OF P.LEIOGNATHI CU,ZN SOD MUTANT V29G | | Descriptor: | COPPER (II) ION, CU,ZN SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Stroppolo, M.E, Pesce, A, D'Orazio, M, O'Neill, P, Bordo, D, Rosano, C, Milani, M, Battistoni, A, Bolognesi, M, Desideri, A. | | Deposit date: | 2001-03-28 | | Release date: | 2001-05-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Single mutations at the subunit interface modulate copper reactivity in Photobacterium leiognathi Cu,Zn superoxide dismutase.
J.Mol.Biol., 308, 2001
|
|
1HTQ
 
 | | Multicopy crystallographic structure of a relaxed glutamine synthetase from Mycobacterium tuberculosis | | Descriptor: | ADENOSINE MONOPHOSPHATE, CITRIC ACID, MANGANESE (II) ION, ... | | Authors: | Gill, H.S, Pfluegl, G.M, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2001-01-01 | | Release date: | 2002-07-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Multicopy crystallographic refinement of a relaxed glutamine synthetase from Mycobacterium tuberculosis highlights flexible loops in the enzymatic mechanism and its regulation.
Biochemistry, 41, 2002
|
|
1SY0
 
 | | 1.15 A Crystal Structure of T121V Mutant of Nitrophorin 4 from Rhodnius Prolixus | | Descriptor: | AMMONIUM ION, Nitrophorin 4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Maes, E.M, Weichsel, A, Andersen, J.F, Shepley, D, Montfort, W.R. | | Deposit date: | 2004-03-31 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Role of binding site loops in controlling nitric oxide release: structure and kinetics of mutant forms of nitrophorin 4
Biochemistry, 43, 2004
|
|
1HMA
 
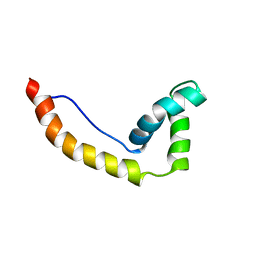 | | THE SOLUTION STRUCTURE AND DYNAMICS OF THE DNA BINDING DOMAIN OF HMG-D FROM DROSOPHILA MELANOGASTER | | Descriptor: | HMG-D | | Authors: | Jones, D.N.M, Searles, M.A, Shaw, G.L, Churchill, M.E.A, Ner, S.S, Keeler, J, Travers, A.A, Neuhaus, D. | | Deposit date: | 1994-05-12 | | Release date: | 1994-07-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure and dynamics of the DNA-binding domain of HMG-D from Drosophila melanogaster.
Structure, 2, 1994
|
|
1HUK
 
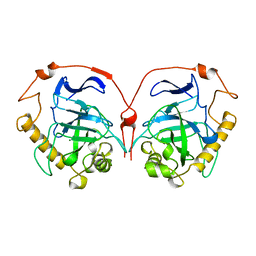 | |
1T2D
 
 | | Plasmodium falciparum lactate dehydrogenase complexed with NAD+ and oxalate | | Descriptor: | GLYCEROL, L-lactate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Cameron, A, Read, J, Tranter, R, Winter, V.J, Sessions, R.B, Brady, R.L, Vivas, L, Easton, A, Kendrick, H, Croft, S.L, Barros, D, Lavandera, J.L, Martin, J.J, Risco, F, Garcia-Ochoa, S, Gamo, F.J, Sanz, L, Leon, L, Ruiz, J.R, Gabarro, R, Mallo, A, De Las Heras, F.G. | | Deposit date: | 2004-04-21 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Identification and Activity of a Series of Azole-based Compounds with Lactate Dehydrogenase-directed Anti-malarial Activity.
J.Biol.Chem., 279, 2004
|
|
1SYS
 
 | | Crystal structure of HLA, B*4403, and peptide EEPTVIKKY | | Descriptor: | Beta-2-microglobulin, Sorting nexin 5, leukocyte antigen (HLA) class I molecule | | Authors: | Zernich, D, Purcell, A.W, Macdonald, W.A, Kjer-Nielsen, L, Ely, L.K, Laham, N, Crockford, T, Mifsud, N.A, Tait, B.D, Holdsworth, R, Brooks, A.G, Bottomley, S.P, Beddoe, T, Peh, C.A, Rossjohn, J, McCluskey, J. | | Deposit date: | 2004-04-01 | | Release date: | 2004-10-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Natural HLA class I polymorphism controls the pathway of antigen presentation and susceptibility to viral evasion
J.Exp.Med., 200, 2004
|
|
2A1T
 
 | | Structure of the human MCAD:ETF E165betaA complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, Acyl-CoA dehydrogenase, medium-chain specific, ... | | Authors: | Toogood, H.S, Van Thiel, A, Scrutton, N.S, Leys, D. | | Deposit date: | 2005-06-21 | | Release date: | 2005-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Stabilization of Non-productive Conformations Underpins Rapid Electron Transfer to Electron-transferring Flavoprotein
J.Biol.Chem., 280, 2005
|
|
1TG3
 
 | | Effect of Shuttle Location and pH Environment on H+ Transfer in Human Carbonic Anhydrase II | | Descriptor: | Carbonic anhydrase II, SULFATE ION, ZINC ION | | Authors: | Fisher, Z, Hernandez Prada, J.A, Tu, C.K, Duda, D, Yoshioka, C, An, H, Govindasamy, L, Silverman, D.N, McKenna, R. | | Deposit date: | 2004-05-28 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Kinetic Characterization of Active-Site Histidine as a Proton Shuttle in Catalysis by Human Carbonic Anhydrase II
Biochemistry, 44, 2005
|
|
2A1X
 
 | | Human phytanoyl-coa 2-hydroxylase in complex with iron and 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, Phytanoyl-CoA dioxygenase | | Authors: | Kavanagh, K.L, McDonough, M.A, Searles, T, Butler, D, Bunkoczi, G, von Delft, F, Edwards, A, Arrowsmith, C, Sundstrom, M, Schofield, C.J, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-06-21 | | Release date: | 2005-08-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Human Phytanoyl-CoA 2-Hydroxylase Identifies Molecular Mechanisms of Refsum Disease
J.Biol.Chem., 280, 2005
|
|
1HK9
 
 | |
