3ZUZ
 
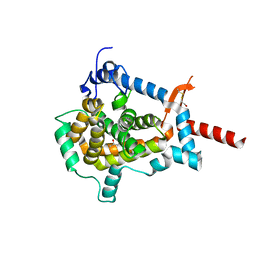 | | Structure of Shq1p C-terminal domain | | Descriptor: | ISOPROPYL ALCOHOL, PROTEIN SHQ1 | | Authors: | Walbott, H, Machado-Pinilla, R, Liger, D, Blaud, M, Rety, S, Grozdanov, P.N, Godin, K, vanTilbeurgh, H, Varani, G, Meier, U.T, Leulliot, N. | | Deposit date: | 2011-07-22 | | Release date: | 2011-11-30 | | Last modified: | 2019-02-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The H/Aca Rnp Assembly Factor Shq1 Functions as an RNA Mimic.
Genes Dev., 25, 2011
|
|
3ZV7
 
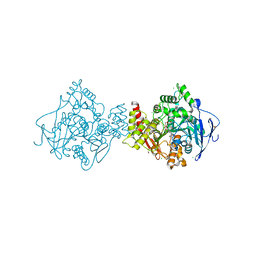 | | Torpedo californica Acetylcholinesterase Inhibition by Bisnorcymserine | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bartolucci, C, Stojan, J, Greig, N.H, Lamba, D. | | Deposit date: | 2011-07-24 | | Release date: | 2012-05-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Kinetics of Torpedo Californica Acetylcholinesterase Inhibition by Bisnorcymserine and Crystal Structure of the Complex with its Leaving Group.
Biochem.J., 444, 2012
|
|
3ZOS
 
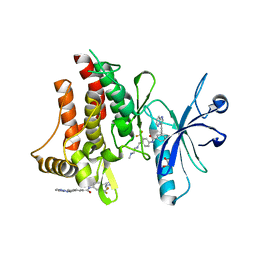 | | Structure of the DDR1 kinase domain in complex with ponatinib | | Descriptor: | 1,2-ETHANEDIOL, 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, EPITHELIAL DISCOIDIN DOMAIN-CONTAINING RECEPTOR 1 | | Authors: | Canning, P, Elkins, J.M, Goubin, S, Mahajan, P, Bradley, A, Coutandin, D, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2013-02-22 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Mechanisms Determining Inhibition of the Collagen Receptor Ddr1 by Selective and Multi-Targeted Type II Kinase Inhibitors
J.Mol.Biol., 426, 2014
|
|
6RRX
 
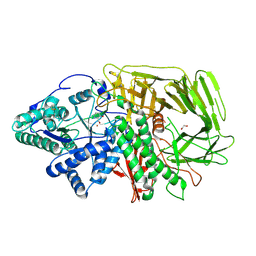 | | GOLGI ALPHA-MANNOSIDASE II in complex with (2S,3R)-2-(Hydroxymethyl)-3-piperidinol | | Descriptor: | (2~{S},3~{R})-2-(hydroxymethyl)piperidin-3-ol, 1,2-ETHANEDIOL, Alpha-mannosidase 2, ... | | Authors: | Armstrong, Z, Lahav, D, Johnson, R, Kuo, C.L, Beenakker, T.J.M, de Boer, C, Wong, C.S, van Rijssel, E.R, Debets, M, Geurink, P.P, Ovaa, H, van der Stelt, M, Codee, J.D.C, Aerts, J.M.F.G, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2019-05-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Manno- epi -cyclophellitols Enable Activity-Based Protein Profiling of Human alpha-Mannosidases and Discovery of New Golgi Mannosidase II Inhibitors.
J.Am.Chem.Soc., 142, 2020
|
|
6RH5
 
 | | Solution structure and 1H, 13C and 15N chemical shift assignments for NECAP1 PHear domain | | Descriptor: | Adaptin ear-binding coat-associated protein 1 | | Authors: | Owen, D.J, Neuhaus, D, Yang, J.-C, Herrmann, T. | | Deposit date: | 2019-04-18 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Temporal Ordering in Endocytic Clathrin-Coated Vesicle Formation via AP2 Phosphorylation.
Dev.Cell, 50, 2019
|
|
3ZRH
 
 | | Crystal structure of the Lys29, Lys33-linkage-specific TRABID OTU deubiquitinase domain reveals an Ankyrin-repeat ubiquitin binding domain (AnkUBD) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, UBIQUITIN THIOESTERASE ZRANB1 | | Authors: | Licchesi, J.D.F, Akutsu, M, Komander, D. | | Deposit date: | 2011-06-16 | | Release date: | 2011-12-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | An Ankyrin-Repeat Ubiquitin-Binding Domain Determines Trabid'S Specificity for Atypical Ubiquitin Chains.
Nat.Struct.Mol.Biol., 19, 2011
|
|
3ZUN
 
 | | pVHL54-213-EloB-EloC complex_(2S,4R)-4-hydroxy-1-(2-(3-methylisoxazol- 5-yl)acetyl)-N-(4-nitrobenzyl)pyrrolidine-2-carboxamide bound | | Descriptor: | (4R)-N-[4-(DIHYDROXYAMINO)BENZYL]-4-HYDROXY-1-[(3-METHYLISOXAZOL-5-YL)ACETYL]-L-PROLINAMIDE, GLYCEROL, TRANSCRIPTION ELONGATION FACTOR B POLYPEPTIDE 1, ... | | Authors: | Van Molle, I, Buckley, D, Crews, C.M, Ciulli, A. | | Deposit date: | 2011-07-19 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Elongin-B, Elongin-C, Von Hippel-Lindau Disease Tumor Suppressor Complex
To be Published
|
|
3ZKE
 
 | | Structure of LC8 in complex with Nek9 peptide | | Descriptor: | DYNEIN LIGHT CHAIN 1, CYTOPLASMIC, NEK9 PROTEIN | | Authors: | Gallego, P, Velazquez-Campoy, A, Regue, L, Roig, J, Reverter, D. | | Deposit date: | 2013-01-22 | | Release date: | 2013-03-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Analysis of the Regulation of the Dynll/Lc8 Binding to Nek9 by Phosphorylation
J.Biol.Chem., 288, 2013
|
|
3ZRA
 
 | | Structural basis for agonism and antagonism for a set of chemically related progesterone receptor modulators | | Descriptor: | N-{(1R)-1-[4-(2-CHLORO-5-FLUOROPYRIDIN-3-YL)PHENYL]ETHYL}-3,5-DIMETHYLISOXAZOLE-4-SULFONAMIDE, PROGESTERONE RECEPTOR, SULFATE ION | | Authors: | Lusher, S.J, Raaijmakers, H.C.A, Vu-Pham, D, Dechering, K, Wai Lam, T, Brown, A.R, Hamilton, N.M, Nimz, O, Azevedo, R, McGuire, R, Oubrie, A, de Vlieg, J. | | Deposit date: | 2011-06-15 | | Release date: | 2011-08-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for agonism and antagonism for a set of chemically related progesterone receptor modulators.
J. Biol. Chem., 286, 2011
|
|
3ZV0
 
 | | Structure of the SHQ1P-CBF5P complex | | Descriptor: | GLYCEROL, H/ACA RIBONUCLEOPROTEIN COMPLEX SUBUNIT 4, PROTEIN SHQ1 | | Authors: | Walbott, H, Machado-Pinilla, R, Liger, D, Blaud, M, Rety, S, Grozdanov, P.N, Godin, K, vanTilbeurgh, H, Varani, G, Meier, U.T, Leulliot, N. | | Deposit date: | 2011-07-22 | | Release date: | 2011-11-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The H/Aca Rnp Assembly Factor Shq1 Functions as an RNA Mimic.
Genes Dev., 25, 2011
|
|
3ZWL
 
 | |
6RRJ
 
 | | GOLGI ALPHA-MANNOSIDASE II in complex with 5-(Adamantan-1yl-methoxy)-pentyl 2,5-dideoxy-2,5-imino-D-talo-hexonamide | | Descriptor: | (2~{S},3~{R},4~{S},5~{R})-~{N}-[5-(1-adamantylmethoxy)pentyl]-5-(hydroxymethyl)-3,4-bis(oxidanyl)pyrrolidine-2-carboxamide, 1,2-ETHANEDIOL, Alpha-mannosidase 2, ... | | Authors: | Armstrong, Z, Lahav, D, Johnson, R, Kuo, C.L, Beenakker, T.J.M, de Boer, C, Wong, C.S, van Rijssel, E.R, Debets, M, Geurink, P.P, Ovaa, H, van der Stelt, M, Codee, J.D.C, Aerts, J.M.F.G, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2019-05-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Manno- epi -cyclophellitols Enable Activity-Based Protein Profiling of Human alpha-Mannosidases and Discovery of New Golgi Mannosidase II Inhibitors.
J.Am.Chem.Soc., 142, 2020
|
|
6S5A
 
 | | CRYSTAL STRUCTURE OF FC P329G LALA WITH ANTI FC P329G FAB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Fc P329G LALA, ... | | Authors: | Ehler, A, Darowski, D, Jost, C, Stubenrauch, K, Wessels, U, Benz, J, Birk, M, Freimoser-Grundschober, A, Bruenker, P, Moessner, E, Umana, P, Kobold, S, Klein, C. | | Deposit date: | 2019-07-01 | | Release date: | 2019-09-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | P329G-CAR-J: a novel Jurkat-NFAT-based CAR-T reporter system recognizing the P329G Fc mutation.
Protein Eng.Des.Sel., 32, 2019
|
|
6RRU
 
 | | GOLGI ALPHA-MANNOSIDASE II in complex with (5R,6R,7S,8S)-5,6,7,8-tetrahydro-5-(hydroxymethyl)-3-(3-phenylpropyl)imidazo[1,2-a]pyridine-6,7,8-triol | | Descriptor: | (5~{R},6~{R},7~{S},8~{S})-5-(hydroxymethyl)-2-(3-phenylpropyl)-5,6,7,8-tetrahydroimidazo[1,2-a]pyridine-6,7,8-triol, Alpha-mannosidase 2, ZINC ION | | Authors: | Armstrong, Z, Lahav, D, Johnson, R, Kuo, C.L, Beenakker, T.J.M, de Boer, C, Wong, C.S, van Rijssel, E.R, Debets, M, Geurink, P.P, Ovaa, H, van der Stelt, M, Codee, J.D.C, Aerts, J.M.F.G, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2019-05-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Manno- epi -cyclophellitols Enable Activity-Based Protein Profiling of Human alpha-Mannosidases and Discovery of New Golgi Mannosidase II Inhibitors.
J.Am.Chem.Soc., 142, 2020
|
|
3ZYH
 
 | | CRYSTAL STRUCTURE OF PA-IL LECTIN COMPLEXED WITH GALBG0 AT 1.5 A RESOLUTION | | Descriptor: | 3-(beta-D-galactopyranosylthio)propanoic acid, CALCIUM ION, PA-I galactophilic lectin | | Authors: | Kadam, R.U, Bergmann, M, Hurley, M, Garg, D, Cacciarini, M, Swiderska, M.A, Nativi, C, Sattler, M, Smyth, A.R, Williams, P, Camara, M, Stocker, A, Darbre, T, Reymond, J.-L. | | Deposit date: | 2011-08-23 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | A Glycopeptide Dendrimer Inhibitor of the Galactose Specific Lectin Leca and of Pseudomonas Aeruginosa Biofilms
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
3ZYB
 
 | | CRYSTAL STRUCTURE OF PA-IL LECTIN COMPLEXED WITH GALAG0 AT 2.3 A RESOLUTION | | Descriptor: | CALCIUM ION, GALA-LYS-PRO-LEUNH2, P-HYDROXYBENZOIC ACID, ... | | Authors: | Kadam, R.U, Bergmann, M, Hurley, M, Garg, D, Cacciarini, M, Swiderska, M.A, Nativi, C, Sattler, M, Smyth, A.R, Williams, P, Camara, M, Stocker, A, Darbre, T, Reymond, J.-L. | | Deposit date: | 2011-08-19 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A Glycopeptide Dendrimer Inhibitor of the Galactose-Specific Lectin Leca and of Pseudomonas Aeruginosa Biofilms.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
3ZYF
 
 | | CRYSTAL STRUCTURE OF PA-IL LECTIN COMPLEXED WITH NPG AT 1.9 A RESOLUTION | | Descriptor: | 4-nitrophenyl beta-D-galactopyranoside, CALCIUM ION, PA-I GALACTOPHILIC LECTIN | | Authors: | Kadam, R.U, Bergmann, M, Hurley, M, Garg, D, Cacciarini, M, Swiderska, M.A, Nativi, C, Sattler, M, Smyth, A.R, Williams, P, Camara, M, Stocker, A, Darbre, T, Reymond, J.-L. | | Deposit date: | 2011-08-22 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.944 Å) | | Cite: | A Glycopeptide Dendrimer Inhibitor of the Galactose-Specific Lectin Leca and of Pseudomonas Aeruginosa Biofilms.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
6RTU
 
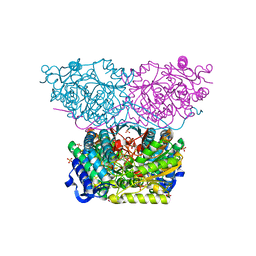 | | Piperideine-6-carboxylate dehydrogenase from Streptomyces clavuligerus complexed with alpha-aminoadipic acid | | Descriptor: | 2-AMINOHEXANEDIOIC ACID, ACETATE ION, GLYCEROL, ... | | Authors: | Hasse, D, Huelsemann, J, Carlsson, G, Andersson, I. | | Deposit date: | 2019-05-26 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of piperideine-6-carboxylate dehydrogenase from Streptomyces clavuligerus.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6RV2
 
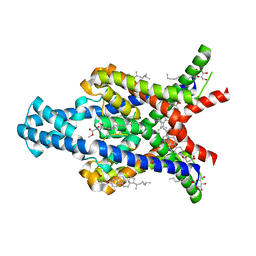 | | Crystal structure of the human two pore domain potassium ion channel TASK-1 (K2P3.1) in a closed conformation | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL HEMISUCCINATE, DECYL-BETA-D-MALTOPYRANOSIDE, ... | | Authors: | Rodstrom, K.E.J, Pike, A.C.W, Zhang, W, Quigley, A, Speedman, D, Mukhopadhyay, S.M.M, Shrestha, L, Chalk, R, Venkaya, S, Bushell, S.R, Tessitore, A, Burgess-Brown, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-05-30 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A lower X-gate in TASK channels traps inhibitors within the vestibule.
Nature, 582, 2020
|
|
6RVS
 
 | | Atomic structure of the Epstein-Barr portal, structure II | | Descriptor: | Portal protein | | Authors: | Machon, C, Fabrega-Ferrer, M, Zhou, D, Cuervo, A, Carrascosa, J.L, Stuart, D.I, Coll, M. | | Deposit date: | 2019-05-31 | | Release date: | 2019-09-18 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Atomic structure of the Epstein-Barr virus portal.
Nat Commun, 10, 2019
|
|
6RNU
 
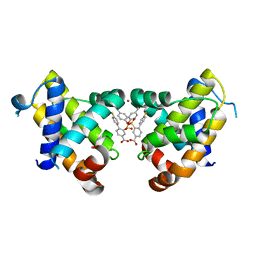 | | BCL-XL in a complex with a covalent small molecule inhibitor | | Descriptor: | 4-(4-fluorophenyl)-3-fluorosulfonyl-benzoic acid, BROMIDE ION, Bcl-2-like protein 1 | | Authors: | Hargreaves, D. | | Deposit date: | 2019-05-09 | | Release date: | 2019-10-02 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery and optimization of covalent Bcl-xL antagonists.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
4BAK
 
 | | Thrombin in complex with inhibitor | | Descriptor: | (2S)-N-(4-CARBAMIMIDOYLBENZYL)-1-[(2R)-2-CYCLOHEXYL-2-{[2-OXO-2-(PROPYLAMINO)ETHYL]AMINO}ACETYL]AZETIDINE-2-CARBOXAMIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, HIRUDIN VARIANT-1, ... | | Authors: | Xue, Y, Musil, D. | | Deposit date: | 2012-09-14 | | Release date: | 2013-01-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Identification of Structure-Kinetic and Structure-Thermodynamic Relationships for Thrombin Inhibitors.
Biochemistry, 52, 2013
|
|
6RPL
 
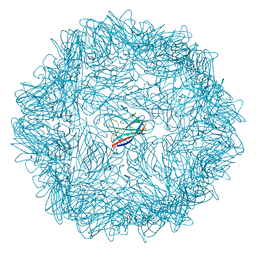 | |
4BB9
 
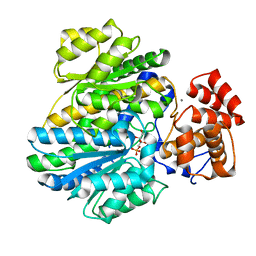 | | Crystal structure of glucokinase regulatory protein complexed to fructose-1-phosphate | | Descriptor: | 1-O-phosphono-beta-D-fructopyranose, CALCIUM ION, GLUCOKINASE REGULATORY PROTEIN | | Authors: | Pautsch, A, Stadler, N, Loehle, A, Lenter, M, Rist, W, Berg, A, Glocker, L, Nar, H, Reinert, D, Heckel, A, Schnapp, G, Kauschke, S.G. | | Deposit date: | 2012-09-21 | | Release date: | 2013-05-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystal Structure of Glucokinase Regulatory Protein.
Biochemistry, 52, 2013
|
|
6RPU
 
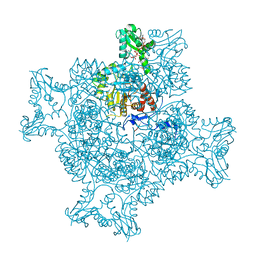 | | Structure of the ternary complex of the IMPDH enzyme from Ashbya gossypii bound to the dinucleoside polyphosphate Ap5G and GDP | | Descriptor: | ACETATE ION, GUANOSINE-5'-DIPHOSPHATE, Inosine-5'-monophosphate dehydrogenase, ... | | Authors: | Buey, R.M, Fernandez-Justel, D, Revuelta, J.L. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The Bateman domain of IMP dehydrogenase is a binding target for dinucleoside polyphosphates.
J.Biol.Chem., 294, 2019
|
|
