6SRV
 
 | | Structure of the arginase-2-inhibitory human antigen-binding fragment Fab C0021144 | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | 著者 | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | 登録日 | 2019-09-06 | | 公開日 | 2020-06-10 | | 最終更新日 | 2020-09-16 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
6SGS
 
 | | Cryo-EM structure of Escherichia coli AcrBZ and DARPin in Saposin A-nanodisc | | 分子名称: | DARPin, Multidrug efflux pump accessory protein AcrZ, Multidrug efflux pump subunit AcrB | | 著者 | Szewczak-Harris, A, Du, D, Newman, C, Neuberger, A, Luisi, B.F. | | 登録日 | 2019-08-05 | | 公開日 | 2020-05-13 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Interactions of a Bacterial RND Transporter with a Transmembrane Small Protein in a Lipid Environment.
Structure, 28, 2020
|
|
6SUQ
 
 | | Crystal Structure of TcdB2-TccC3-TEV | | 分子名称: | TcdB2,TccC3,Genome polyprotein | | 著者 | Roderer, D, Schubert, E, Sitsel, O, Raunser, S. | | 登録日 | 2019-09-16 | | 公開日 | 2019-12-04 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Towards the application of Tc toxins as a universal protein translocation system.
Nat Commun, 10, 2019
|
|
7F55
 
 | | Cryo-EM structure of bremelanotide-MC4R-Gs_Nb35 complex | | 分子名称: | CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Zhang, H, Chen, L, Mao, C, Shen, Q, Yang, D, Shen, D, Qin, J. | | 登録日 | 2021-06-21 | | 公開日 | 2021-11-03 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural insights into ligand recognition and activation of the melanocortin-4 receptor.
Cell Res., 31, 2021
|
|
3EKB
 
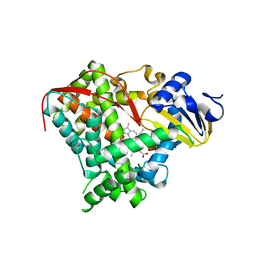 | |
7F53
 
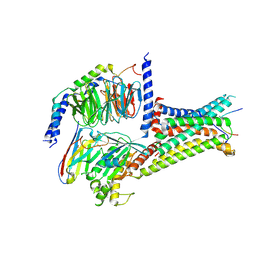 | | Cryo-EM structure of a-MSH-MC4R-Gs_Nb35 complex | | 分子名称: | CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Zhang, H, Chen, L, Mao, C, Shen, Q, Yang, D, Shen, D, Qin, J. | | 登録日 | 2021-06-21 | | 公開日 | 2021-11-03 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural insights into ligand recognition and activation of the melanocortin-4 receptor.
Cell Res., 31, 2021
|
|
6SX5
 
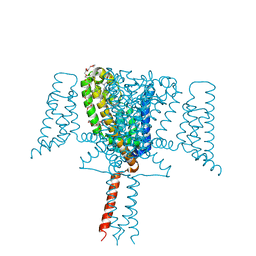 | | Full length Mutant Open-form Sodium Channel NavMs | | 分子名称: | DODECAETHYLENE GLYCOL, HEGA-10, Ion transport protein, ... | | 著者 | Sula, A, Hollingworth, D, Wallace, B.A. | | 登録日 | 2019-09-25 | | 公開日 | 2021-02-03 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | A tamoxifen receptor within a voltage-gated sodium channel.
Mol.Cell, 81, 2021
|
|
6SXC
 
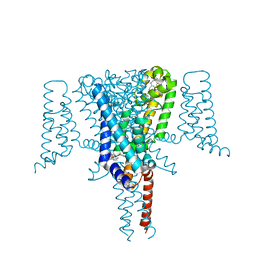 | | Crystal structure of the Voltage-Gated Sodium Channel NavMs (F208L) in complex with 4-hydroxytamoxifen (2.5 Angstrom resolution) | | 分子名称: | 4-HYDROXYTAMOXIFEN, DODECAETHYLENE GLYCOL, HEGA-10, ... | | 著者 | Sula, A, Hollingworth, D, Wallace, B.A. | | 登録日 | 2019-09-25 | | 公開日 | 2021-02-03 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A tamoxifen receptor within a voltage-gated sodium channel.
Mol.Cell, 81, 2021
|
|
6SPC
 
 | | Pseudomonas aeruginosa 30s ribosome from an aminoglycoside resistant clinical isolate | | 分子名称: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | 著者 | Halfon, Y, Jimenez-Fernande, A, La Ros, R, Espinos, R, Krogh Johansen, H, Matzov, D, Eyal, Z, Bashan, A, Zimmerman, E, Belousoff, M, Molin, S, Yonath, A. | | 登録日 | 2019-09-01 | | 公開日 | 2019-10-16 | | 最終更新日 | 2019-11-06 | | 実験手法 | ELECTRON MICROSCOPY (2.95 Å) | | 主引用文献 | Structure ofPseudomonas aeruginosaribosomes from an aminoglycoside-resistant clinical isolate.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6SYZ
 
 | | Crystal structure of YTHDC1 with fragment 1 (DHU_DC1_141) | | 分子名称: | SULFATE ION, YTH domain-containing protein 1, ~{N}-methylthieno[3,2-d]pyrimidin-4-amine | | 著者 | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | 登録日 | 2019-10-01 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
6SRU
 
 | | Structure of Ig-like V-type domian of mouse Programmed cell death 1 ligand 1 (PD-L1) | | 分子名称: | Programmed cell death 1 ligand 1 | | 著者 | Magiera-Mularz, K, Sala, D, Grudnik, P, Holak, T.A. | | 登録日 | 2019-09-06 | | 公開日 | 2021-02-03 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.532 Å) | | 主引用文献 | Human and mouse PD-L1: similar molecular structure, but different druggability profiles.
Iscience, 24, 2021
|
|
6T01
 
 | | Crystal structure of YTHDC1 with fragment 13 (DHU_DC1_153) | | 分子名称: | 6-methyl-5-nitro-4-phenyl-1~{H}-pyrimidin-2-one, SULFATE ION, YTH domain-containing protein 1 | | 著者 | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | 登録日 | 2019-10-02 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
6T0A
 
 | | Crystal structure of YTHDC1 with fragment 25 (PSI_DC1_005) | | 分子名称: | (~{R})-azanyl(pyridin-3-yl)methanethiol, SULFATE ION, YTHDC1 | | 著者 | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | 登録日 | 2019-10-02 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
3E9N
 
 | | Crystal structure of a putative short-chain dehydrogenase/reductase from Corynebacterium glutamicum | | 分子名称: | PUTATIVE SHORT-CHAIN DEHYDROGENASE/REDUCTASE | | 著者 | Bonanno, J.B, Gilmore, M, Bain, K.T, Hu, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2008-08-22 | | 公開日 | 2008-09-02 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of a putative short-chain dehydrogenase/reductase from Corynebacterium glutamicum
To be Published
|
|
6SXF
 
 | | Crystal Structure of the Voltage-Gated Sodium Channel NavMs (F208L) in complex with Tamoxifen (2.8 Angstrom resolution) | | 分子名称: | (Z)-2-[4-(1,2)-DIPHENYL-1-BUTENYL)-PHENOXY]-N,N-DIMETHYLETHANAMINE, DODECAETHYLENE GLYCOL, HEGA-10, ... | | 著者 | Sula, A, Hollingworth, D, Wallace, B.A. | | 登録日 | 2019-09-25 | | 公開日 | 2021-02-03 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.839 Å) | | 主引用文献 | A tamoxifen receptor within a voltage-gated sodium channel.
Mol.Cell, 81, 2021
|
|
6SZP
 
 | | High resolution crystal structure of human DDAH-1 in complex with N-(4-Aminobutyl)-N'-(2-Methoxyethyl)guanidine | | 分子名称: | (1~{S})-~{N}'-(4-azanylbutyl)-~{N}"-(2-methoxyethyl)methanetriamine, GLYCEROL, N(G),N(G)-dimethylarginine dimethylaminohydrolase 1 | | 著者 | Hennig, S, Vetter, I.R, Schade, D. | | 登録日 | 2019-10-02 | | 公開日 | 2019-12-25 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Discovery ofN-(4-Aminobutyl)-N'-(2-methoxyethyl)guanidine as the First Selective, Nonamino Acid, Catalytic Site Inhibitor of Human Dimethylarginine Dimethylaminohydrolase-1 (hDDAH-1).
J.Med.Chem., 63, 2020
|
|
3EIV
 
 | |
6SL5
 
 | | Dunaliella Photosystem I Supercomplex | | 分子名称: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | 著者 | Nelson, N, Caspy, I, Malavath, T, Klaiman, D, Shkolinsky, Y. | | 登録日 | 2019-08-18 | | 公開日 | 2020-06-24 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (2.84 Å) | | 主引用文献 | Structure and energy transfer pathways of the Dunaliella Salina photosystem I supercomplex.
Biochim Biophys Acta Bioenerg, 1861, 2020
|
|
3EDZ
 
 | | Crystal structure of catalytic domain of TACE with hydroxamate inhibitor | | 分子名称: | ADAM 17, CITRIC ACID, N-{(2R)-2-[2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}-3-methyl-L-valyl-N-(2-aminoethyl)-L-alaninamide, ... | | 著者 | Mazzola, R.D, Zhu, Z, Sinning, L, McKittrick, B, Lavey, B, Spitler, J, Kozlowski, J, Neng-Yang, S, Zhou, G, Guo, Z, Orth, P, Madison, V, Sun, J, Lundell, D, Niu, X. | | 登録日 | 2008-09-03 | | 公開日 | 2008-09-23 | | 最終更新日 | 2021-10-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Discovery of novel hydroxamates as highly potent tumor necrosis factor-alpha converting enzyme inhibitors. Part II: optimization of the S3' pocket.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
6SS3
 
 | | Kemp Eliminase HG3.17 mutant Q50K, E47N, N300D Complexed with Transition State Analog 6-Nitrobenzotriazole | | 分子名称: | 6-NITROBENZOTRIAZOLE, GLYCEROL, Kemp Eliminase HG3.17 Q50K, ... | | 著者 | Bloch, J.S, Pinkas, D.M, Hilvert, D. | | 登録日 | 2019-09-06 | | 公開日 | 2020-04-01 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Contribution of Oxyanion Stabilization to Kemp Eliminase Efficiencyproficiency
Acs Catalysis, 2020
|
|
6SRY
 
 | | Kemp Eliminase HG3.17 mutant Q50S, E47N, N300D Complexed with Transition State Analog 6-Nitrobenzotriazole | | 分子名称: | 6-NITROBENZOTRIAZOLE, Kemp Eliminase HG3.17 Q50S, E47N,N300D, ... | | 著者 | Bloch, J.S, Pinkas, D.M, Hilvert, D. | | 登録日 | 2019-09-06 | | 公開日 | 2020-04-01 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (0.95 Å) | | 主引用文献 | Contribution of Oxyanion Stabilization to Kemp Eliminase Efficiencyproficiency
Acs Catalysis, 2020
|
|
3E5Z
 
 | | X-Ray structure of the putative gluconolactonase in protein family PF08450. Northeast Structural Genomics Consortium target DrR130. | | 分子名称: | MAGNESIUM ION, putative Gluconolactonase | | 著者 | Kuzin, A.P, Abashidze, M, Seetharaman, J, Wang, D, Mao, L, Maglaqui, M, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Tong, S.N, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2008-08-14 | | 公開日 | 2008-09-30 | | 最終更新日 | 2017-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | X-Ray structure of the putative gluconolactonase in protein family PF08450. Northeast Structural Genomics Consortium target DrR130.
To be Published
|
|
6STA
 
 | | Crystal structure of the strawberry pathogenesis-related 10 (PR-10) Fra a 1.02 protein, E46A D48A mutant | | 分子名称: | Major strawberry allergen Fra a 1-2 | | 著者 | Orozco-Navarrete, B, Kaczmarska, Z, Dupeux, F, Pott, D, Diaz Perales, A, Casanal, A, Marquez, J.A, Valpuesta, V, Merchante, C. | | 登録日 | 2019-09-10 | | 公開日 | 2019-12-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Structural Bases for the Allergenicity of Fra a 1.02 in Strawberry Fruits.
J.Agric.Food Chem., 68, 2020
|
|
3E8T
 
 | | Crystal Structure of Epiphyas postvittana Takeout 1 | | 分子名称: | Takeout-like protein 1, Ubiquinone-8 | | 著者 | Hamiaux, C, Stanley, D, Greenwood, D.R, Baker, E.N, Newcomb, R.D. | | 登録日 | 2008-08-20 | | 公開日 | 2008-12-09 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Crystal structure of Epiphyas postvittana takeout 1 with bound ubiquinone supports a role as ligand carriers for takeout proteins in insects
J.Biol.Chem., 284, 2009
|
|
6ST9
 
 | | Crystal structure of the strawberry pathogenesis-related 10 (PR-10) Fra a 1.02 protein, D48R mutant | | 分子名称: | CHLORIDE ION, Major strawberry allergen Fra a 1-2 | | 著者 | Orozco-Navarrete, B, Kaczmarska, Z, Dupeux, F, Pott, D, Diaz Perales, A, Casanal, A, Marquez, J.A, Valpuesta, V, Merchante, C. | | 登録日 | 2019-09-10 | | 公開日 | 2019-12-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Structural Bases for the Allergenicity of Fra a 1.02 in Strawberry Fruits.
J.Agric.Food Chem., 68, 2020
|
|
