6S4F
 
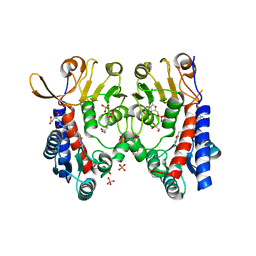 | | Structure of human MTHFD2 in complex with TH9619 | | Descriptor: | (E,4S)-4-[[5-[2-[2,6-bis(azanyl)-4-oxidanylidene-1H-pyrimidin-5-yl]ethanoylamino]-3-fluoranyl-pyridin-2-yl]carbonylamino]pent-2-enedioic acid, ADENOSINE-5'-DIPHOSPHATE, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, ... | | Authors: | Scaletti, E.R, Gustafsson, R, Bonagas, N, Gustafsson, N.M, Henriksson, M, Abdurakhmanov, E, Andersson, Y, Bengtsson, C, Borhade, S, Desroses, M, Farnegardh, K, Garg, N, Gokturk, C, Haraldsson, M, Iliev, P, Jarvius, M, Jemth, A.S, Kalderen, C, Karsten, S, Klingegard, F, Koolmeister, T, Martens, U, Llona-Minguez, S, Loseva, O, Marttila, P, Michel, M, Moulson, R, Nordstrom, H, Paulin, C, Pham, T, Pudelko, L, Rasti, A, Roos, A.K, Sarno, A, Sandberg, L, Scobie, M, Sjoberg, B, Svensson, R, Unterlass, J.E, Vallin, K, Vo, D, Wiita, E, Warpman-Berglund, U, Homan, E.J, Helleday, T, Stenmark, P. | | Deposit date: | 2019-06-27 | | Release date: | 2021-07-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Pharmacological targeting of MTHFD2 suppresses acute myeloid leukemia by inducing thymidine depletion and replication stress.
Nat Cancer, 3, 2022
|
|
6S4E
 
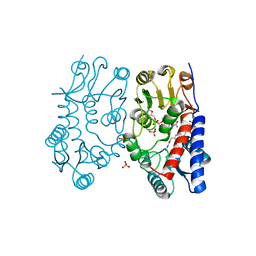 | | Structure of human MTHFD2 in complex with TH7299 | | Descriptor: | (2S)-2-[[4-[[2,4-bis(azanyl)-6-oxidanylidene-1H-pyrimidin-5-yl]carbamoylamino]phenyl]carbonylamino]pentanedioic acid, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, mitochondrial, ... | | Authors: | Gustafsson, R, Scaletti, E.R, Bonagas, N, Gustafsson, N.M, Henriksson, M, Abdurakhmanov, E, Andersson, Y, Bengtsson, C, Borhade, S, Desroses, M, Farnegardh, K, Garg, N, Gokturk, C, Haraldsson, M, Iliev, P, Jarvius, M, Jemth, A.S, Kalderen, C, Karsten, S, Klingegard, F, Koolmeister, T, Martens, U, Llona-Minguez, S, Loseva, O, Marttila, P, Michel, M, Moulson, R, Nordstrom, H, Paulin, C, Pham, T, Pudelko, L, Rasti, A, Roos, A.K, Sarno, A, Sandberg, L, Scobie, M, Sjoberg, B, Svensson, R, Unterlass, J.E, Vallin, K, Vo, D, Wiita, E, Warpman-Berglund, U, Homan, E.J, Helleday, T, Stenmark, P. | | Deposit date: | 2019-06-27 | | Release date: | 2021-07-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pharmacological targeting of MTHFD2 suppresses acute myeloid leukemia by inducing thymidine depletion and replication stress.
Nat Cancer, 3, 2022
|
|
3ZM7
 
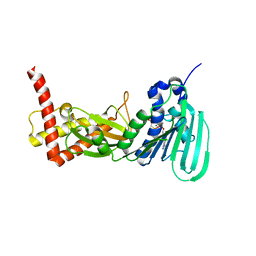 | | CRYSTAL STRUCTURE OF THE ATPASE REGION OF Mycobacterium tuberculosis GyrB WITH AMPPCP | | Descriptor: | DNA GYRASE SUBUNIT B, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Agrawal, A, Roue, M, Spitzfaden, C, Petrella, S, Aubry, A, Volker, C, Mossakowska, D, Hann, M, Bax, B, Mayer, C. | | Deposit date: | 2013-02-05 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Mycobacterium Tuberculosis DNA Gyrase ATPase Domain Structures Suggest a Dissociative Mechanism that Explains How ATP Hydrolysis is Coupled to Domain Motion.
Biochem.J., 456, 2013
|
|
3HSV
 
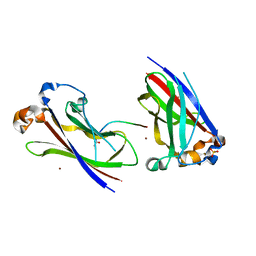 | | Structures of SPOP-Substrate Complexes: Insights into Molecular Architectures of BTB-Cul3 Ubiquitin Ligases: SPOPMATHx-MacroH2ASBCpep2 | | Descriptor: | Core histone macro-H2A.1, SULFATE ION, Speckle-type POZ protein, ... | | Authors: | Zhuang, M, Schulman, B.A, Miller, D. | | Deposit date: | 2009-06-10 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structures of SPOP-substrate complexes: insights into molecular architectures of BTB-Cul3 ubiquitin ligases.
Mol.Cell, 36, 2009
|
|
2OJW
 
 | | Crystal structure of human glutamine synthetase in complex with ADP and phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Karlberg, T, Uppenberg, J, Arrowsmith, C, Berglund, H, Busam, R.D, Collins, R, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hallberg, B.M, Hammarstrom, M, Hogbom, M, Johansson, I, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Van Den Berg, S, Wallden, K, Weigelt, J, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-15 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of mammalian glutamine synthetases illustrate substrate-induced conformational changes and provide opportunities for drug and herbicide design.
J.Mol.Biol., 375, 2008
|
|
6S8W
 
 | | Aromatic aminotransferase AroH (Aro8) form Aspergillus fumigatus in complex with PLP (internal aldimine) | | Descriptor: | Aromatic aminotransferase Aro8, putative, FORMIC ACID, ... | | Authors: | Giardina, G, Mirco, D, Spizzichino, S, Zelante, T, Cutruzzola, F, Romani, L, Cellini, B. | | Deposit date: | 2019-07-10 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Aspergillus fumigatus AroH, an aromatic amino acid aminotransferase
Proteins, 2021
|
|
2NUK
 
 | | Soluble Domain of the Rieske Iron-Sulfur Protein from Rhodobacter sphaeroides | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Ubiquinol-cytochrome c reductase iron-sulfur subunit | | Authors: | Kolling, D, Brunzelle, J, Lhee, S, Crofts, A.R, Nair, S.K. | | Deposit date: | 2006-11-09 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Atomic resolution structures of rieske iron-sulfur protein: role of hydrogen bonds in tuning the redox potential of iron-sulfur clusters.
Structure, 15, 2007
|
|
2NVF
 
 | | Soluble domain of Rieske Iron-Sulfur protein. | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, Ubiquinol-cytochrome c reductase iron-sulfur subunit | | Authors: | Kolling, D, Brunzelle, J, Lhee, S, Crofts, A.R, Nair, S.K. | | Deposit date: | 2006-11-12 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Atomic resolution structures of rieske iron-sulfur protein: role of hydrogen bonds in tuning the redox potential of iron-sulfur clusters.
Structure, 15, 2007
|
|
2NWF
 
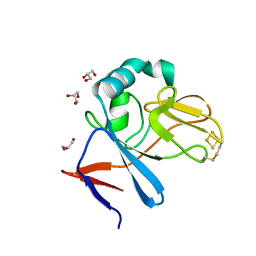 | | Soluble domain of Rieske Iron Sulfur Protein | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, Ubiquinol-cytochrome c reductase iron-sulfur subunit | | Authors: | Kolling, D, Brunzelle, J.S, Lhee, S, Crofts, A.R, Nair, S.K. | | Deposit date: | 2006-11-14 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic resolution structures of rieske iron-sulfur protein: role of hydrogen bonds in tuning the redox potential of iron-sulfur clusters.
Structure, 15, 2007
|
|
6SHT
 
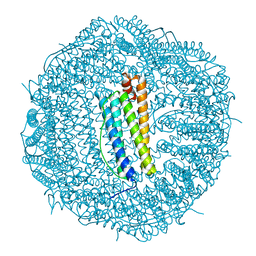 | | Molecular structure of mouse apoferritin resolved at 2.7 Angstroms with the Glacios cryo-microscope | | Descriptor: | FE (III) ION, Ferritin heavy chain, MAGNESIUM ION | | Authors: | Hamdi, F, Tueting, C, Semchonok, D, Kyrilis, F, Meister, A, Skalidis, I, Schmidt, L, Parthier, C, Stubbs, M.T, Kastritis, P.L. | | Deposit date: | 2019-08-08 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | 2.7 angstrom cryo-EM structure of vitrified M. musculus H-chain apoferritin from a compact 200 keV cryo-microscope.
Plos One, 15, 2020
|
|
3IQM
 
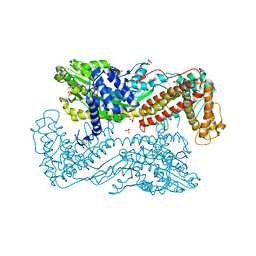 | | Active site mutants of B. subtilis SecA | | Descriptor: | Protein translocase subunit secA, SULFATE ION | | Authors: | Kim, D, Hunt, J.F. | | Deposit date: | 2009-08-20 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | ATPase Active-Site Electrostatic Interactions Control the Global Conformation of the 100 kDa SecA Translocase.
J.Am.Chem.Soc., 135, 2013
|
|
3IVN
 
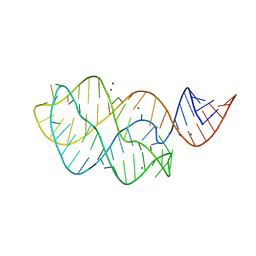 | | Structure of the U65C mutant A-riboswitch aptamer from the Bacillus subtilis pbuE operon | | Descriptor: | A-riboswitch, BROMIDE ION, MAGNESIUM ION | | Authors: | Delfosse, V, Dagenais, P, Chausse, D, Di Tomasso, G, Legault, P. | | Deposit date: | 2009-09-01 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Riboswitch structure: an internal residue mimicking the purine ligand.
Nucleic Acids Res., 38, 2010
|
|
6SGF
 
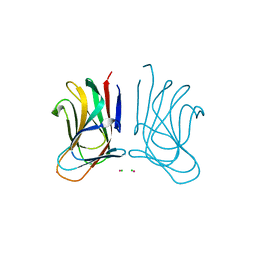 | | Molecular insight into a new low affinity xylan binding module CBM86, from the xylanolytic gut symbiont Roseburia intestinalis. | | Descriptor: | Beta-xylanase, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Ejby, M, Abou Hachem, M, Leth, M.L, Guskov, A, Slotboom, D. | | Deposit date: | 2019-08-04 | | Release date: | 2019-11-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.756 Å) | | Cite: | Molecular insight into a new low-affinity xylan binding module from the xylanolytic gut symbiont Roseburia intestinalis.
Febs J., 287, 2020
|
|
2OT3
 
 | |
3ZKB
 
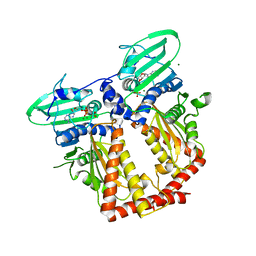 | | CRYSTAL STRUCTURE OF THE ATPASE REGION OF Mycobacterium tuberculosis GyrB WITH AMPPNP | | Descriptor: | DNA GYRASE SUBUNIT B, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Agrawal, A, Roue, M, Spitzfaden, C, Petrella, S, Aubry, A, Volker, C, Mossakowska, D, Hann, M, Bax, B, Mayer, C. | | Deposit date: | 2013-01-22 | | Release date: | 2013-09-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mycobacterium Tuberculosis DNA Gyrase ATPase Domain Structures Suggest a Dissociative Mechanism that Explains How ATP Hydrolysis is Coupled to Domain Motion.
Biochem.J., 456, 2013
|
|
3J04
 
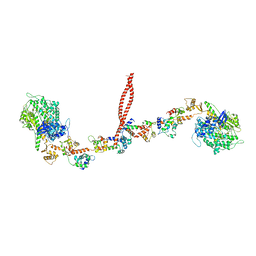 | | EM structure of the heavy meromyosin subfragment of Chick smooth muscle Myosin with regulatory light chain in phosphorylated state | | Descriptor: | Myosin light polypeptide 6, Myosin regulatory light chain 2, smooth muscle major isoform, ... | | Authors: | Baumann, B.A.J, Taylor, D, Huang, Z, Tama, F, Fagnant, P.M, Trybus, K, Taylor, K. | | Deposit date: | 2011-02-18 | | Release date: | 2011-11-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Phosphorylated smooth muscle heavy meromyosin shows an open conformation linked to activation.
J.Mol.Biol., 415, 2012
|
|
3L0F
 
 | |
4B2M
 
 | | COMPLEXES OF DODECIN WITH FLAVIN AND FLAVIN-LIKE LIGANDS | | Descriptor: | CHLORIDE ION, DODECIN, MAGNESIUM ION, ... | | Authors: | Staudt, H, Hoesl, M, Dreuw, A, Serdjukow, S, Oesterhelt, D, Budisa, N, Wachtveitl, J, Grininger, M. | | Deposit date: | 2012-07-16 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The flavoprotein dodecin as a redox probe for electron transfer through DNA.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
3LDY
 
 | | An extraordinary mechanism of DNA perturbation exhibited by the rare-cutting HNH restriction endonuclease PacI | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*AP*GP*GP*CP*TP*TP*A)-3'), DNA (5'-D(P*AP*TP*TP*AP*AP*GP*CP*CP*TP*C)-3'), ... | | Authors: | Shen, B.W, Heiter, D, Chan, S.-H, Xu, S.-Y, Wilson, G, Stoddard, B.L. | | Deposit date: | 2010-01-13 | | Release date: | 2010-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Unusual target site disruption by the rare-cutting HNH restriction endonuclease PacI.
Structure, 18, 2010
|
|
2PSO
 
 | | Human StarD13 (DLC2) lipid transfer and protein localization domain | | Descriptor: | StAR-related lipid transfer protein 13 | | Authors: | Lehtio, L, Busam, R, Arrowsmith, C.H, Berglund, H, Collins, R, Dahlgren, L.G, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg-Schiavone, L, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Moche, M, Nordlund, P, Nyman, T, Ogg, D, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Van den Berg, S, Weigelt, J, Welin, M, Persson, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-07 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Comparative structural analysis of lipid binding START domains.
Plos One, 6, 2011
|
|
3ZHF
 
 | | gamma 2 adaptin EAR domain crystal structure with preS1 site1 peptide NPDWDFN | | Descriptor: | 1,2-ETHANEDIOL, AP-1 COMPLEX SUBUNIT GAMMA-LIKE 2, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Juergens, M.C, Voros, J, Rautureau, G, Shepherd, D, Pye, V.E, Muldoon, J, Johnson, C.M, Ashcroft, A, Freund, S.M.V, Ferguson, N. | | Deposit date: | 2012-12-21 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Hepatitis B Virus Pres1 Domain Hijacks Host Trafficking Proteins by Motif Mimicry.
Nat.Chem.Biol., 9, 2013
|
|
2PMQ
 
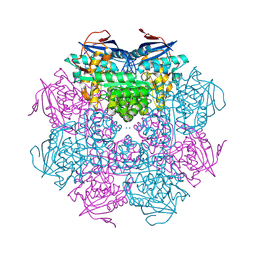 | | Crystal structure of a mandelate racemase/muconate lactonizing enzyme from Roseovarius sp. HTCC2601 | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Lau, C, Sridhar, V, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-04-23 | | Release date: | 2007-05-08 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of new enzymes and metabolic pathways by using structure and genome context.
Nature, 502, 2013
|
|
3LH9
 
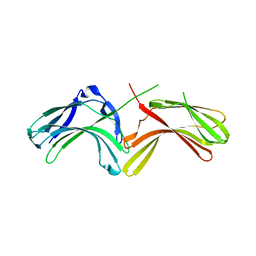 | |
4BCX
 
 | | gamma 2 adaptin EAR domain crystal structure | | Descriptor: | 1,3-PROPANDIOL, AP-1 COMPLEX SUBUNIT GAMMA-LIKE 2, IMIDAZOLE | | Authors: | Juergens, M.C, Voros, J, Rautureau, G, Shepherd, D, Pye, V.E, Muldoon, J, Johnson, C.M, Ashcroft, A, Freund, S.M.V, Ferguson, N. | | Deposit date: | 2012-10-03 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Hepatitis B Virus Pres1 Domain Hijacks Host Trafficking Proteins by Motif Mimicry.
Nat.Chem.Biol., 9, 2013
|
|
3LH8
 
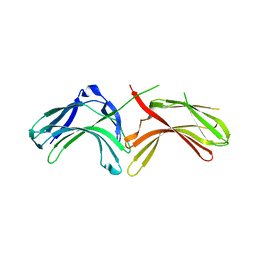 | |
