3GA0
 
 | | CtBP1/BARS Gly172->Glu mutant structure: impairing NAD(H) binding and dimerization | | Descriptor: | C-terminal-binding protein 1, FORMIC ACID | | Authors: | Nardini, M, Valente, C, Ricagno, S, Luini, A, Corda, D, Bolognesi, M. | | Deposit date: | 2009-02-16 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | CtBP1/BARS Gly172-->Glu mutant structure: impairing NAD(H)-binding and dimerization
Biochem.Biophys.Res.Commun., 381, 2009
|
|
3GAC
 
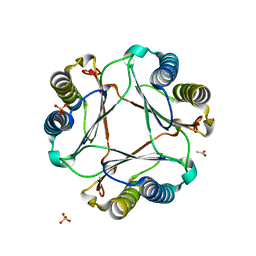 | | Structure of mif with HPP | | Descriptor: | 3-(4-HYDROXY-PHENYL)PYRUVIC ACID, ACETIC ACID, Macrophage migration inhibitory factor-like protein, ... | | Authors: | Zhou, Y.-F, Su, X.-D, Shao, D, Wang, H. | | Deposit date: | 2009-02-17 | | Release date: | 2009-12-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional comparison of MIF ortholog from Plasmodium yoelii with MIF from its rodent host
Mol.Immunol., 47, 2010
|
|
6TUB
 
 | | Beta-endorphin amyloid fibril | | Descriptor: | Beta-endorphin | | Authors: | Verasdonck, J, Seuring, C, Gath, J, Ghosh, D, Nespovitaya, N, Waelti, M.A, Maji, S, Cadalbert, R, Boeckmann, A, Guentert, P, Meier, B.H, Riek, R. | | Deposit date: | 2020-01-05 | | Release date: | 2020-10-28 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | The three-dimensional structure of human beta-endorphin amyloid fibrils.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6T5J
 
 | | Structure of NUDT15 in complex with inhibitor TH1760 | | Descriptor: | 6-[4-(1~{H}-indol-5-ylcarbonyl)piperazin-1-yl]sulfonyl-3~{H}-1,3-benzoxazol-2-one, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Carter, M, Rehling, D, Desroses, M, Zhang, S.M, Hagenkort, A, Valerie, N.C.K, Helleday, T, Stenmark, P. | | Deposit date: | 2019-10-16 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Development of a chemical probe against NUDT15.
Nat.Chem.Biol., 16, 2020
|
|
6TFY
 
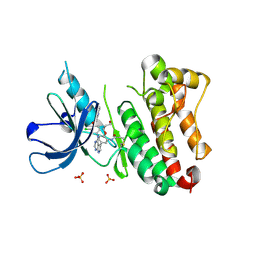 | | Crystal Structure of EGFR T790M/V948R in Complex with Covalent Pyrrolopyrimidine 18c | | Descriptor: | Epidermal growth factor receptor, SULFATE ION, ~{N}-[5-[4-[[3-chloranyl-4-(pyridin-2-ylmethoxy)phenyl]amino]-7~{H}-pyrrolo[2,3-d]pyrimidin-5-yl]-2-(3-oxidanylpropoxy)phenyl]propanamide | | Authors: | Niggenaber, J, Mueller, M.P, Rauh, D. | | Deposit date: | 2019-11-14 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Targeting Her2-insYVMA with Covalent Inhibitors-A Focused Compound Screening and Structure-Based Design Approach.
J.Med.Chem., 63, 2020
|
|
3G6S
 
 | | Crystal structure of the endonuclease/exonuclease/phosphatase (BVU_0621) from Bacteroides vulgatus. Northeast Structural Genomics Consortium Target BvR56D | | Descriptor: | Putative endonuclease/exonuclease/phosphatase family protein | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Xiao, R, Sahdev, S, Foote, E.L, Ciccosanti, C, Wang, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-02-08 | | Release date: | 2009-02-17 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the endonuclease/exonuclease/phosphatase (BVU_0621) from Bacteroides vulgatus.
To be Published
|
|
6TTV
 
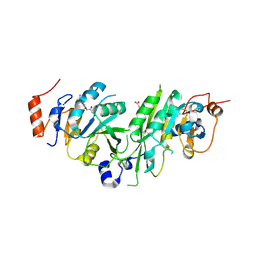 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 3 (ASI_M3M_138) | | Descriptor: | (2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolane-2-carboxamide, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-12-30 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Small-Molecule Inhibitors of METTL3, the Major Human Epitranscriptomic Writer.
Chemmedchem, 15, 2020
|
|
6THE
 
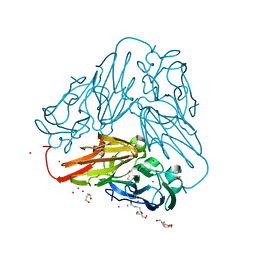 | | Crystal structure of core domain of four-domain heme-cupredoxin-Cu nitrite reductase from Bradyrhizobium sp. ORS 375 | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Sasaki, D, Watanabe, T.F, Eady, R.R, Garratt, R.C, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2019-11-20 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Reverse protein engineering of a novel 4-domain copper nitrite reductase reveals functional regulation by protein-protein interaction.
Febs J., 288, 2021
|
|
6TUI
 
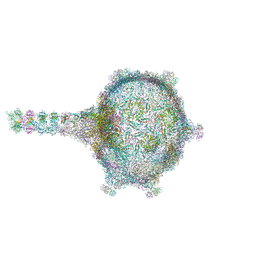 | | Virion of empty GTA particle | | Descriptor: | Adaptor protein Rcc01688, Phage major capsid protein, HK97 family, ... | | Authors: | Bardy, P, Fuzik, T, Hrebik, D, Pantucek, R, Beatty, J.T, Plevka, P. | | Deposit date: | 2020-01-07 | | Release date: | 2020-10-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (10.47 Å) | | Cite: | Virion of empty GTA particle
Nat Commun, 11, 2020
|
|
6T1N
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with benzimidazole-amide derivative 5 | | Descriptor: | 1,2-ETHANEDIOL, 4-chloranyl-~{N}-[2-(piperidin-1-ylmethyl)-3~{H}-benzimidazol-5-yl]benzamide, Protein ENL | | Authors: | Chaikuad, A, Heidenreich, D, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
3GA2
 
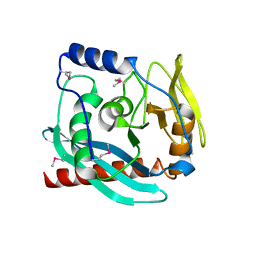 | | Crystal structure of the Endonuclease_V (BSU36170) from Bacillus subtilis, Northeast Structural Genomics Consortium Target SR624 | | Descriptor: | Endonuclease V | | Authors: | Forouhar, F, Abashidze, M, Hussain, M, Seetharaman, J, Janjua, H, Fang, Y, Xiao, R, Cunningham, K, Ma, L.-C, Owens, L, Wang, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-02-16 | | Release date: | 2009-02-24 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the Endonuclease_V (BSU36170) from Bacillus subtilis, Northeast Structural Genomics Consortium Target SR624
To be Published
|
|
6TM5
 
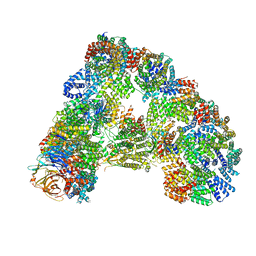 | |
6T4H
 
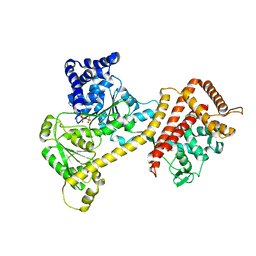 | | Crystal structure of the accessory translocation ATPase, SecA2, from Clostridium difficile, in complex with adenosine-5'-(gamma-thio)-triphosphate | | Descriptor: | PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Protein translocase subunit SecA 2 | | Authors: | Lindic, N, Loboda, J, Usenik, A, Turk, D. | | Deposit date: | 2019-10-14 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Structure of Clostridioides difficile SecA2 ATPase Exposes Regions Responsible for Differential Target Recognition of the SecA1 and SecA2-Dependent Systems.
Int J Mol Sci, 21, 2020
|
|
6T3B
 
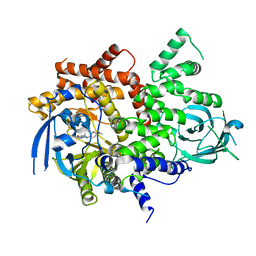 | | Crystal structure of PI3Kgamma with a dihydropurinone inhibitor (compound 4) | | Descriptor: | 2-[(4-methoxy-2-methyl-phenyl)amino]-7-methyl-9-(4-oxidanylcyclohexyl)purin-8-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Petersen, J, Oster, L, Schimpl, M, Goldberg, F.W, Finlay, M.R.V, Ting, A.K.T, Beattie, D, Lamont, G.M, Fallan, C, Wrigley, G.L, Howard, M.R, Williamson, B, Davies, B.R, Cadogan, E.B, Ramos-Montoya, A, Dean, E. | | Deposit date: | 2019-10-10 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | The Discovery of 7-Methyl-2-[(7-methyl[1,2,4]triazolo[1,5-a]pyridin-6-yl)amino]-9-(tetrahydro-2H-pyran-4-yl)-7,9-dihydro-8H-purin-8-one (AZD7648), a Potent and Selective DNA-Dependent Protein Kinase (DNA-PK) Inhibitor.
J.Med.Chem., 63, 2020
|
|
3FWJ
 
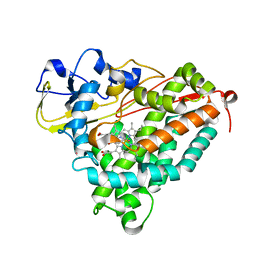 | | Ferric camphor bound Cytochrome P450cam containing a selenocysteine as the 5th heme ligand, orthorombic crystal form | | Descriptor: | CAMPHOR, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Schlichting, I, von Koenig, K, Aldag, C, Hilvert, D. | | Deposit date: | 2009-01-18 | | Release date: | 2009-03-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the role of the proximal heme ligand in cytochrome P450cam by recombinant incorporation of selenocysteine.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6THF
 
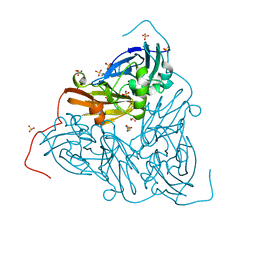 | | Crystal structure of two-domain Cu nitrite reductase from Bradyrhizobium sp. ORS 375 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Sasaki, D, Watanabe, T.F, Eady, R.R, Garratt, R.C, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2019-11-20 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Reverse protein engineering of a novel 4-domain copper nitrite reductase reveals functional regulation by protein-protein interaction.
Febs J., 288, 2021
|
|
6T6B
 
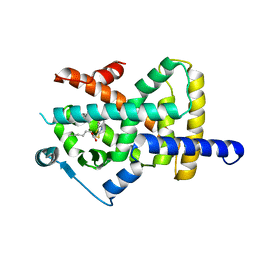 | | Crystal structure of PPARgamma in complex with compound 16 (MF27) | | Descriptor: | (2~{R})-2-[[6-[(2,4-dichlorophenyl)sulfonylamino]-1,3-benzothiazol-2-yl]sulfanyl]octanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Chaikuad, A, Ni, X, Hanke, T, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-18 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Selective Modulator of Peroxisome Proliferator-Activated Receptor gamma with an Unprecedented Binding Mode.
J.Med.Chem., 63, 2020
|
|
6T1O
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with benzimidazole-amide derivative 6 | | Descriptor: | 1,2-ETHANEDIOL, 4-iodanyl-~{N}-[2-(piperidin-1-ylmethyl)-3~{H}-benzimidazol-5-yl]benzamide, Protein ENL | | Authors: | Chaikuad, A, Heidenreich, D, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
3G9K
 
 | | Crystal structure of Bacillus anthracis transpeptidase enzyme CapD | | Descriptor: | Capsule biosynthesis protein capD, GLUTAMIC ACID | | Authors: | Zhang, R, Wu, R, Richter, S, Anderson, V.J, Missiakas, D, Joachimiak, A. | | Deposit date: | 2009-02-13 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal Structure of Bacillus anthracis Transpeptidase Enzyme CapD.
J.Biol.Chem., 284, 2009
|
|
3GBV
 
 | | Crystal structure of a putative LacI transcriptional regulator from Bacteroides fragilis | | Descriptor: | 1,2-ETHANEDIOL, Putative LacI-family transcriptional regulator, SODIUM ION | | Authors: | Syed Ibrahim, B, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-20 | | Release date: | 2009-03-10 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a putative LacI transcriptional regulator from Bacteroides fragilis
To be Published
|
|
6TA0
 
 | | Human NAMPT in complex with nicotinic acid and phosphoribosyl pyrophosphate | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, GLYCEROL, NICOTINIC ACID, ... | | Authors: | Houry, D, Raasakka, A, Kursula, P, Ziegler, M. | | Deposit date: | 2019-10-29 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Identification of structural determinants of NAMPT activity and substrate selectivity
To Be Published
|
|
6TKH
 
 | | Tsetse thrombin inhibitor in complex with human alpha-thrombin - orthorhombic form at 7keV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SODIUM ION, ... | | Authors: | Calisto, B.M, Ripoll-Rozada, J, de Sanctis, D, Pereira, P.J.B. | | Deposit date: | 2019-11-28 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sulfotyrosine-Mediated Recognition of Human Thrombin by a Tsetse Fly Anticoagulant Mimics Physiological Substrates.
Cell Chem Biol, 28, 2021
|
|
3GJ2
 
 | | Photoactivated state of PA-GFP | | Descriptor: | CHLORIDE ION, Green fluorescent protein | | Authors: | Henderson, J.N, Gepshtein, R, Heenan, J.R, Kallio, K, Huppert, D, Remington, S.J. | | Deposit date: | 2009-03-07 | | Release date: | 2009-03-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of the photoactivatable green fluorescent protein.
J.Am.Chem.Soc., 131, 2009
|
|
3GCU
 
 | | Human P38 MAP kinase in complex with RL48 | | Descriptor: | 1-{3-[(6-aminoquinazolin-4-yl)amino]phenyl}-3-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]urea, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mitogen-activated protein kinase 14, ... | | Authors: | Gruetter, C, Simard, J.R, Getlik, M, Rauh, D. | | Deposit date: | 2009-02-22 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Development of a fluorescent-tagged kinase assay system for the detection and characterization of allosteric kinase inhibitors.
J.Am.Chem.Soc., 131, 2009
|
|
6UCT
 
 | | Crystal structure of Mal de Rio Cuarto virus P9-1 viroplasm protein (C-arm deletion mutant) | | Descriptor: | p9-1 | | Authors: | Llauger, G, Klinke, S, Monti, D, Sycz, G, Cerutti, M.L, Goldbaum, F.A, del Vas, M, Otero, L.H. | | Deposit date: | 2019-09-17 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | Crystal structure of Mal de Rio Cuarto virus P9-1 viroplasm protein (C-arm deletion mutant)
To Be Published
|
|
