4N9W
 
 | | Crystal structure of phosphatidyl mannosyltransferase PimA | | Descriptor: | 1,2-ETHANEDIOL, GDP-mannose-dependent alpha-(1-2)-phosphatidylinositol mannosyltransferase, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Giganti, D, Albesa-Jove, D, Bellinzoni, M, Guerin, M.E, Alzari, P.M. | | Deposit date: | 2013-10-21 | | Release date: | 2014-11-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Secondary structure reshuffling modulates glycosyltransferase function at the membrane.
Nat.Chem.Biol., 11, 2015
|
|
4TNE
 
 | | Crystal Structure of Human Transthyretin Thr119Tyr Mutant | | Descriptor: | GLYCEROL, Transthyretin | | Authors: | Saelices, L, Cascio, D, Sawaya, M, Eisenberg, D.S. | | Deposit date: | 2014-06-03 | | Release date: | 2015-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Uncovering the Mechanism of Aggregation of Human Transthyretin.
J.Biol.Chem., 290, 2015
|
|
4TPV
 
 | | Crystal Structure of Hookworm Platelet Inhibitor | | Descriptor: | GLYCEROL, Platelet inhibitor | | Authors: | Andersen, J.F, Ma, D, Francischetti, I. | | Deposit date: | 2014-06-09 | | Release date: | 2015-04-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of hookworm platelet inhibitor (HPI), a CAP superfamily member from Ancylostoma caninum.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
7SJO
 
 | | HtrA1S328A:Fab15H6.v4 complex | | Descriptor: | Fab15H6.v4 Heavy Chain, Fab15H6.v4 Light Chain, Serine protease HTRA1 | | Authors: | Gerhardy, S, Green, E, Estevez, A, Arthur, C.P, Ultsch, M, Rohou, A, Kirchhofer, D. | | Deposit date: | 2021-10-18 | | Release date: | 2022-09-07 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Allosteric inhibition of HTRA1 activity by a conformational lock mechanism to treat age-related macular degeneration.
Nat Commun, 13, 2022
|
|
7N0H
 
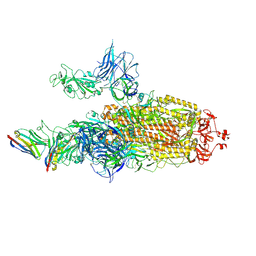 | | CryoEM structure of SARS-CoV-2 spike protein (S-6P, 2-up) in complex with sybodies (Sb45) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Jiang, J, Huang, R, Margulies, D. | | Deposit date: | 2021-05-25 | | Release date: | 2021-06-02 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
7N0G
 
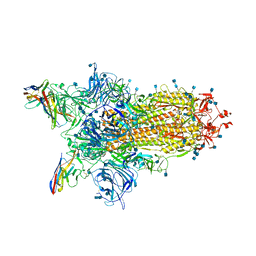 | | CryoEm structure of SARS-CoV-2 spike protein (S-6P, 1-up) in complex with sybodies (Sb45) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Jiang, J, Huang, R, Margulies, D. | | Deposit date: | 2021-05-25 | | Release date: | 2021-06-02 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
4S1L
 
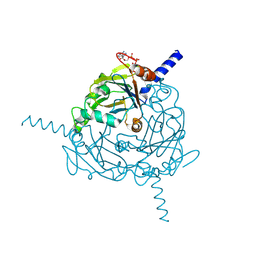 | | Structure of Uranotaenia sapphirina cypovirus (CPV17) polyhedrin at 298 K | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, polyhedrin | | Authors: | Ginn, H.M, Messerschmidt, M, Ji, X, Zhang, H, Axford, D, Gildea, R.J, Winter, G, Brewster, A.S, Hattne, J, Wagner, A, Grimes, J.M, Evans, G, Sauter, N.K, Sutton, G, Stuart, D.I. | | Deposit date: | 2015-01-14 | | Release date: | 2015-03-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Structure of CPV17 polyhedrin determined by the improved analysis of serial femtosecond crystallographic data.
Nat Commun, 6, 2015
|
|
7SF3
 
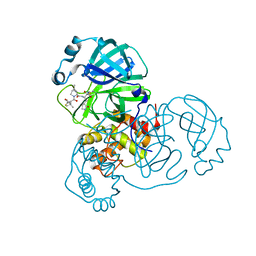 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML1006m | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-3-hydroxy-4-(methylamino)-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase, CHLORIDE ION | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2021-10-02 | | Release date: | 2022-10-05 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
4TLS
 
 | | Crystal Structure of Human Transthyretin Glu92Pro Mutant | | Descriptor: | GLYCEROL, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Saelices, L, Cascio, D, Sawaya, M, Eisenberg, D.S. | | Deposit date: | 2014-05-30 | | Release date: | 2015-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Uncovering the Mechanism of Aggregation of Human Transthyretin.
J.Biol.Chem., 290, 2015
|
|
7SFH
 
 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML102 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-(3-phenylpropanoyl)-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase, CALCIUM ION | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2021-10-03 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Rational design of a new class of protease inhibitors for the potential treatment of coronavirus diseases
To Be Published
|
|
5VR0
 
 | |
7SFB
 
 | |
7SGH
 
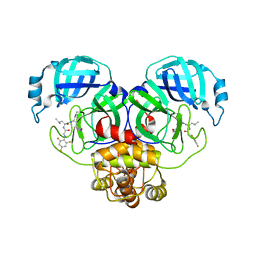 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML124N | | Descriptor: | (S)-N-((S)-1-imino-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)-4-methyl-2-(2-((2,4,6-trifluorophenyl)amino)acetamido)pentanamide, 3C-like proteinase | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2021-10-05 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Rational design of a new class of protease inhibitors for the potential treatment of coronavirus diseases
To Be Published
|
|
7SFI
 
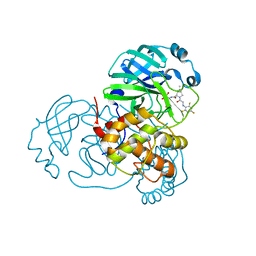 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML104 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-[N-(2,4,6-trifluorophenyl)glycyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase, CALCIUM ION, ... | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2021-10-03 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Rational design of a new class of protease inhibitors for the potential treatment of coronavirus diseases
To Be Published
|
|
4TR2
 
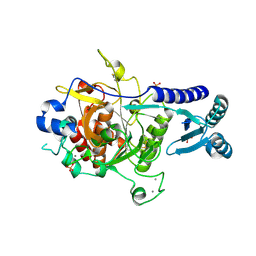 | | Crystal structure of PvSUB1 | | Descriptor: | CALCIUM ION, PHOSPHATE ION, Subtilisin-like 1 serine protease | | Authors: | Giganti, D, Bouillon, A, Martinez, M, Weber, P, Girard-Blanc, C, Petres, S, Haouz, A, Barale, J.C, Alzari, P.M. | | Deposit date: | 2014-06-13 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A novel Plasmodium-specific prodomain fold regulates the malaria drug target SUB1 subtilase.
Nat Commun, 5, 2014
|
|
4TRQ
 
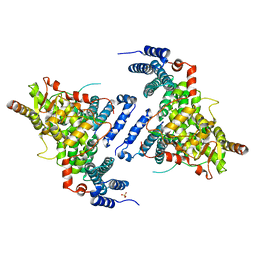 | | Crystal structure of Sac3/Thp1/Sem1 | | Descriptor: | 26S proteasome complex subunit SEM1, Nuclear mRNA export protein SAC3, Nuclear mRNA export protein THP1, ... | | Authors: | Hellerschmied, D, Schneider, S, Kohler, A, Clausen, T. | | Deposit date: | 2014-06-17 | | Release date: | 2015-08-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Nuclear Pore-Associated TREX-2 Complex Employs Mediator to Regulate Gene Expression.
Cell, 162, 2015
|
|
1KH0
 
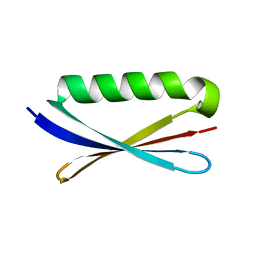 | | Accurate Computer Base Design of a New Backbone Conformation in the Second Turn of Protein L | | Descriptor: | protein L | | Authors: | O'Neill, J.W, Kuhlman, B, Kim, D.E, Zhang, K.Y, Baker, D. | | Deposit date: | 2001-11-28 | | Release date: | 2002-01-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Accurate computer-based design of a new backbone conformation in the second turn of protein L.
J.Mol.Biol., 315, 2002
|
|
8ENX
 
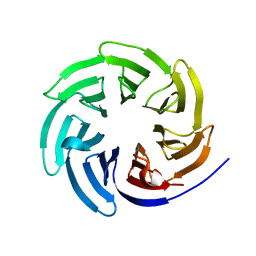 | |
8ENY
 
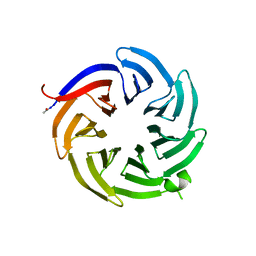 | |
8ENZ
 
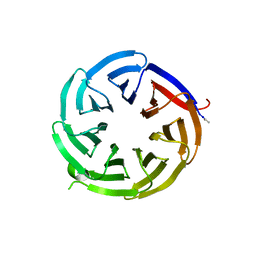 | |
8EO0
 
 | |
4TKH
 
 | | The 0.93 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with myristic acid | | Descriptor: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Matsuoka, S, Mizohata, E, Matsuoka, D, Ishida, H, Hirose, M, Kakinouchi, K, Hara, T, Murakami, S, Inoue, T, Murata, M. | | Deposit date: | 2014-05-26 | | Release date: | 2015-01-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | Water-mediated recognition of simple alkyl chains by heart-type Fatty-Acid-binding protein
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
8ENS
 
 | |
7SET
 
 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML1000 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase, CHLORIDE ION | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2021-10-01 | | Release date: | 2022-10-05 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
8ENW
 
 | |
