2ZMA
 
 | | Crystal Structure of 6-Aminohexanoate-dimer Hydrolase S112A/G181D/H266N/D370Y Mutant with Substrate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Ohki, T, Shibata, N, Higuchi, Y, Takeo, M, Negoro, S. | | Deposit date: | 2008-04-14 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Molecular design of a nylon-6 byproduct-degrading enzyme from a carboxylesterase with a beta-lactamase fold
Febs J., 276, 2009
|
|
2ZM7
 
 | | Structure of 6-Aminohexanoate-dimer Hydrolase, S112A/G181D Mutant Complexed with 6-Aminohexanoate-dimer | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Ohki, T, Shibata, N, Higuchi, Y, Kawashima, Y, Takeo, M, Kato, D, Negoro, S. | | Deposit date: | 2008-04-14 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular design of a nylon-6 byproduct-degrading enzyme from a carboxylesterase with a beta-lactamase fold
Febs J., 276, 2009
|
|
1RCP
 
 | | CYTOCHROME C' | | Descriptor: | CYTOCHROME C', PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tahirov, T.H, Misaki, S, Meyer, T.E, Cusanovich, M.A, Higuchi, Y, Yasuoka, N. | | Deposit date: | 1995-08-23 | | Release date: | 1996-06-10 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution crystal structures of two polymorphs of cytochrome c' from the purple phototrophic bacterium rhodobacter capsulatus.
J.Mol.Biol., 259, 1996
|
|
3WIH
 
 | | Crystal structure of the third fibronectin domain (Fn3) of human ROBO1 in complex with the Fab fragment of murine monoclonal antibody B2212A. | | Descriptor: | GLYCEROL, Roundabout homolog 1, anti-human ROBO1 antibody B2212A Fab heavy chain, ... | | Authors: | Nakayama, T, Mizohata, E, Yamashita, T, Nagatoishi, M, Iwanari, H, Mochizuki, Y, Kado, Y, Yokota, Y, Sato, R, Tsumoto, K, Fujitani, H, Kodama, T, Hamakubo, T, Inoue, T. | | Deposit date: | 2013-09-12 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Structural features of interfacial tyrosine residue in ROBO1 fibronectin domain-antibody complex: Crystallographic, thermodynamic, and molecular dynamic analyses
Protein Sci., 24, 2015
|
|
3WKV
 
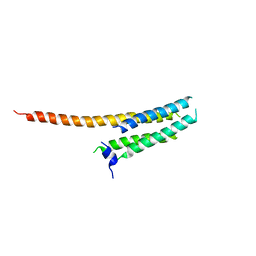 | | Voltage-gated proton channel: VSOP/Hv1 chimeric channel | | Descriptor: | Ion channel | | Authors: | Takeshita, K, Sakata, S, Yamashita, E, Fujiwara, Y, Kawanabe, A, Kurokawa, T, Okochi, Y, Matsuda, M, Narita, H, Okamura, Y, Nakagawa, A. | | Deposit date: | 2013-10-31 | | Release date: | 2014-03-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.453 Å) | | Cite: | X-ray crystal structure of voltage-gated proton channel.
Nat.Struct.Mol.Biol., 21, 2014
|
|
3A66
 
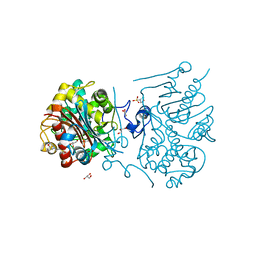 | | Crystal structure of 6-aminohexanoate-dimer hydrolase S112A/G181D/H266N/D370Y mutant with substrate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOATE-DIMER HYDROLASE, 6-AMINOHEXANOIC ACID, ... | | Authors: | Kawashima, Y, Shibata, N, Higuchi, Y, Takeo, M, Negoro, S. | | Deposit date: | 2009-08-21 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Enzymatic Synthesis of Nylon-6 Units in Organic Sol Contained Low-Water: Structural Requirement of 6-Aminohexanoate-Dimer Hydrolase for Efficient Amid Synthesis
To be Published
|
|
3A65
 
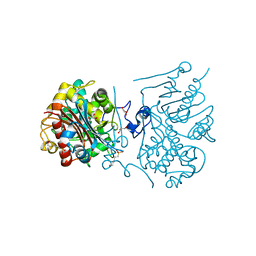 | | Crystal structure of 6-aminohexanoate-dimer hydrolase S112A/G181D/H266N mutant with substrate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Kawashima, Y, Shibata, N, Higuchi, Y, Takeo, M, Negoro, S. | | Deposit date: | 2009-08-21 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Enzymatic Synthesis of Nylon-6 Units in Organic Sol Contained Low-Water: Structural Requirement of 6-Aminohexanoate-Dimer Hydrolase for Efficient Amid Synthesis
To be Published
|
|
3VR1
 
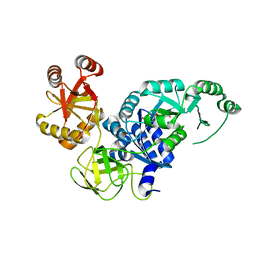 | | Crystal structure analysis of the translation factor RF3 | | Descriptor: | GUANOSINE-5',3'-TETRAPHOSPHATE, Peptide chain release factor 3 | | Authors: | Kihira, K, Shomura, Y, Shibata, N, Kitamura, M, Higuchi, Y. | | Deposit date: | 2012-04-03 | | Release date: | 2012-09-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure analysis of the translation factor RF3 (release factor 3)
Febs Lett., 586, 2012
|
|
2KCX
 
 | |
7BQV
 
 | | Cereblon in complex with SALL4 and (S)-5-hydroxythalidomide | | Descriptor: | 2-[(3~{S})-2,6-bis(oxidanylidene)piperidin-3-yl]-5-oxidanyl-isoindole-1,3-dione, Protein cereblon, SULFATE ION, ... | | Authors: | Furihata, H, Miyauchi, Y, Asano, A, Tanokura, M, Miyakawa, T. | | Deposit date: | 2020-03-25 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural bases of IMiD selectivity that emerges by 5-hydroxythalidomide.
Nat Commun, 11, 2020
|
|
5GZ8
 
 | |
7BQU
 
 | | Cereblon in complex with SALL4 and (S)-thalidomide | | Descriptor: | Protein cereblon, S-Thalidomide, Sal-like protein 4, ... | | Authors: | Furihata, H, Miyauchi, Y, Asano, A, Tanokura, M, Miyakawa, T. | | Deposit date: | 2020-03-25 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural bases of IMiD selectivity that emerges by 5-hydroxythalidomide.
Nat Commun, 11, 2020
|
|
8ZMG
 
 | | Crystal structure of an inverse agonist antipsychotic drug pimavanserin-bound 5-HT2A | | Descriptor: | 5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, Pimavanserin | | Authors: | Oguma, T, Asada, H, Sekiguchi, Y, Imono, M, Iwata, S, Kusakabe, K. | | Deposit date: | 2024-05-23 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Dual 5-HT 2A and 5-HT 2C Receptor Inverse Agonist That Affords In Vivo Antipsychotic Efficacy with Minimal hERG Inhibition for the Treatment of Dementia-Related Psychosis.
J.Med.Chem., 2024
|
|
3VHQ
 
 | | Crystal structure of the Ca6 site mutant of Pro-SA-subtilisin | | Descriptor: | CALCIUM ION, Tk-subtilisin | | Authors: | Uehara, R, Takeuchi, Y, Tanaka, S, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2011-09-01 | | Release date: | 2012-07-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Requirement of Ca(2+) Ions for the Hyperthermostability of Tk-Subtilisin from Thermococcus kodakarensis
Biochemistry, 51, 2012
|
|
3VTH
 
 | |
3VYM
 
 | | Dimeric Hydrogenobacter thermophilus cytochrome c552 | | Descriptor: | Cytochrome c-552, HEME C | | Authors: | Hayashi, Y, Nagao, S, Osuka, H, Komori, H, Higuchi, Y, Hirota, S. | | Deposit date: | 2012-09-28 | | Release date: | 2012-11-07 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Domain Swapping of the Heme and N-Terminal alpha-Helix in Hydrogenobacter thermophilus Cytochrome c(552) Dimer
Biochemistry, 51, 2012
|
|
3VTI
 
 | | Crystal structure of HypE-HypF complex | | Descriptor: | FE (III) ION, Hydrogenase maturation factor, MAGNESIUM ION, ... | | Authors: | Shomura, Y, Higuchi, Y. | | Deposit date: | 2012-05-30 | | Release date: | 2012-07-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural basis for the reaction mechanism of S-carbamoylation of HypE by HypF in the maturation of [NiFe]-hydrogenases
J.Biol.Chem., 287, 2012
|
|
3VUZ
 
 | | Crystal structure of histone methyltransferase SET7/9 in complex with AAM-1 | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl](hexyl)amino}-5'-deoxyadenosine, Histone-lysine N-methyltransferase SETD7 | | Authors: | Niwa, H, Handa, N, Tomabechi, Y, Honda, K, Toyama, M, Ohsawa, N, Shirouzu, M, Kagechika, H, Hirano, T, Umehara, T, Yokoyama, S. | | Deposit date: | 2012-07-10 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of histone methyltransferase SET7/9 in complexes with adenosylmethionine derivatives
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2KL7
 
 | |
6IQY
 
 | | High resolution structure of bilirubin oxidase from Myrothecium verrucaria - M467Q mutant, anaerobically prepared | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Bilirubin oxidase, ... | | Authors: | Shibata, N, Akter, M, Higuchi, Y. | | Deposit date: | 2018-11-09 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Redox Potential-Dependent Formation of an Unusual His-Trp Bond in Bilirubin Oxidase.
Chemistry, 24, 2018
|
|
3VV0
 
 | | Crystal structure of histone methyltransferase SET7/9 in complex with DAAM-3 | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl][2-(hexylamino)ethyl]amino}-5'-deoxyadenosine, Histone-lysine N-methyltransferase SETD7 | | Authors: | Niwa, H, Handa, N, Tomabechi, Y, Honda, K, Toyama, M, Ohsawa, N, Shirouzu, M, Kagechika, H, Hirano, T, Umehara, T, Yokoyama, S. | | Deposit date: | 2012-07-10 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structures of histone methyltransferase SET7/9 in complexes with adenosylmethionine derivatives
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5GZ9
 
 | | Crystal structure of catalytic domain of Protein O-mannosyl Kinase in complexes with AMP-PNP, Magnesium ions and glycopeptide | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Protein O-mannose kinase, ... | | Authors: | Nagae, M, Yamaguchi, Y. | | Deposit date: | 2016-09-27 | | Release date: | 2017-03-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 3D structural analysis of protein O-mannosyl kinase, POMK, a causative gene product of dystroglycanopathy.
Genes Cells, 22, 2017
|
|
5GYR
 
 | | Tetrameric Allochromatium vinosum cytochrome c' | | Descriptor: | Cytochrome c', HEME C | | Authors: | Yamanaka, M, Hoshizumi, M, Nagao, S, Nakayama, R, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2016-09-23 | | Release date: | 2017-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Formation and carbon monoxide-dependent dissociation of Allochromatium vinosum cytochrome c' oligomers using domain-swapped dimers
Protein Sci., 26, 2017
|
|
6IQX
 
 | | High resolution structure of bilirubin oxidase from Myrothecium verrucaria - M467Q mutant, aerobically prepared | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Bilirubin oxidase, ... | | Authors: | Shibata, N, Akter, M, Higuchi, Y. | | Deposit date: | 2018-11-09 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.432 Å) | | Cite: | Redox Potential-Dependent Formation of an Unusual His-Trp Bond in Bilirubin Oxidase.
Chemistry, 24, 2018
|
|
6IQZ
 
 | | High resolution structure of bilirubin oxidase from Myrothecium verrucaria - wild type | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bilirubin oxidase, COPPER (II) ION, ... | | Authors: | Shibata, N, Akter, M, Higuchi, Y. | | Deposit date: | 2018-11-09 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Redox Potential-Dependent Formation of an Unusual His-Trp Bond in Bilirubin Oxidase.
Chemistry, 24, 2018
|
|
