5KQJ
 
 | |
6CEY
 
 | | Aminoglycoside Phosphotransferase (2'')-Ia in complex with GMPPNP, Magnesium, and Lividomycin moieties | | Descriptor: | (2R,3S,4S,5S,6R)-2-((2S,3S,4R,5R,6R)-5-AMINO-2-(AMINOMETHYL)-6-((2R,3S,4R,5S)-5-((1R,2R,3S,5R,6S)-3,5-DIAMINO-2-((2S,3R ,5S,6R)-3-AMINO-5-HYDROXY-6-(HYDROXYMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-6-HYDROXYCYCLOHEXYLOXY)-4-HYDROXY-2-(HYDROXYMET HYL)-TETRAHYDROFURAN-3-YLOXY)-4-HYDROXY-TETRAHYDRO-2H-PYRAN-3-YLOXY)-6-(HYDROXYMETHYL)-TETRAHYDRO-2H-PYRAN-3,4,5-TRIOL, Bifunctional AAC/APH, CHLORIDE ION, ... | | Authors: | Caldwell, S.J, Berghuis, A.M. | | Deposit date: | 2018-02-13 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Plasticity of Aminoglycoside Binding to Antibiotic Kinase APH(2′′)-Ia.
Antimicrob. Agents Chemother., 62, 2018
|
|
1BWD
 
 | | INOSAMINE-PHOSPHATE AMIDINOTRANSFERASE STRB1 FROM STREPTOMYCES GRISEUS | | Descriptor: | PROTEIN (INOSAMINE-PHOSPHATE AMIDINOTRANSFERASE) | | Authors: | Fritsche, E, Bergner, A, Humm, A, Piepersberg, W, Huber, R. | | Deposit date: | 1998-09-23 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of L-arginine:inosamine-phosphate amidinotransferase StrB1 from Streptomyces griseus: an enzyme involved in streptomycin biosynthesis.
Biochemistry, 37, 1998
|
|
7LQY
 
 | | Structure of squirrel TRPV1 in apo state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoinositol, CHLORIDE ION, ... | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2021-02-15 | | Release date: | 2021-04-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Extracellular cap domain is an essential component of the TRPV1 gating mechanism.
Nat Commun, 12, 2021
|
|
1914
 
 | | SIGNAL RECOGNITION PARTICLE ALU RNA BINDING HETERODIMER, SRP9/14 | | Descriptor: | BETA-MERCAPTOETHANOL, PHOSPHATE ION, SIGNAL RECOGNITION PARTICLE 9/14 FUSION PROTEIN | | Authors: | Birse, D, Kapp, U, Strub, K, Cusack, S, Aberg, A. | | Deposit date: | 1997-11-13 | | Release date: | 1998-12-30 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | The crystal structure of the signal recognition particle Alu RNA binding heterodimer, SRP9/14.
EMBO J., 16, 1997
|
|
4QXS
 
 | | Crystal structure of human FPPS in complex with WC01088 | | Descriptor: | (2-{2-[(2S)-3-methylbutan-2-yl]-5-phenyl-1H-indol-3-yl}ethane-1,1-diyl)bis(phosphonic acid), Farnesyl pyrophosphate synthase, GLYCEROL, ... | | Authors: | Park, J, Zielinski, M, Weiling, C, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2014-07-21 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the molecular and structural elements of ligands binding to the active site versus an allosteric pocket of the human farnesyl pyrophosphate synthase.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
1GZ9
 
 | | High-Resolution Crystal Structure of Erythrina cristagalli Lectin in Complex with 2'-alpha-L-Fucosyllactose | | Descriptor: | CALCIUM ION, ERYTHRINA CRISTA-GALLI LECTIN, MANGANESE (II) ION, ... | | Authors: | Svensson, C, Teneberg, S, Nilsson, C.L, Kjellberg, A, Schwarz, F.P, Sharon, N, Krengel, U. | | Deposit date: | 2002-05-17 | | Release date: | 2002-06-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-Resolution Crystal Structures of Erythrina Cristagalli Lectin in Complex with Lactose and 2'-Alpha-L-Fucosyllactose and Correlation with Thermodynamic Binding Data
J.Mol.Biol., 321, 2002
|
|
6CGD
 
 | | Aminoglycoside Phosphotransferase (2'')-Ia in complex with GMPPNP, Magnesium, and Amikacin | | Descriptor: | (2S)-N-[(1R,2S,3S,4R,5S)-4-[(2R,3R,4S,5S,6R)-6-(aminomethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-5-azanyl-2-[(2S,3R,4S,5S ,6R)-4-azanyl-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]oxy-3-oxidanyl-cyclohexyl]-4-azanyl-2-oxidanyl-butanamide, Bifunctional AAC/APH, CHLORIDE ION, ... | | Authors: | Caldwell, S.J, Berghuis, A.M. | | Deposit date: | 2018-02-20 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Plasticity of Aminoglycoside Binding to Antibiotic Kinase APH(2′′)-Ia.
Antimicrob. Agents Chemother., 62, 2018
|
|
4XJE
 
 | |
4ZS7
 
 | | Structural mimicry of receptor interaction by antagonistic IL-6 antibodies | | Descriptor: | Interleukin-6, Llama Fab fragment 68F2 heavy chain, Llama Fab fragment 68F2 light chain | | Authors: | Blanchetot, C, De Jonge, N, Desmyter, A, Ongenae, N, Hofman, E, Klarenbeek, A, Sadi, A, Hultberg, A, Kretz-Rommel, A, Spinelli, S, Loris, R, Cambillau, C, de Haard, H. | | Deposit date: | 2015-05-13 | | Release date: | 2016-05-04 | | Last modified: | 2016-07-06 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Structural Mimicry of Receptor Interaction by Antagonistic Interleukin-6 (IL-6) Antibodies.
J.Biol.Chem., 291, 2016
|
|
6C57
 
 | | Crystal structure of mutant human geranylgeranyl pyrophosphate synthase (Y246D) in complex with bisphosphonate inhibitor FV0109 | | Descriptor: | Geranylgeranyl pyrophosphate synthase, {[(2-{3-[(4-fluorobenzene-1-carbonyl)amino]phenyl}thieno[2,3-d]pyrimidin-4-yl)amino]methylene}bis(phosphonic acid) | | Authors: | Park, J, Bin, X, Vincent, F, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2018-01-15 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Unraveling the Prenylation-Cancer Paradox in Multiple Myeloma with Novel Geranylgeranyl Pyrophosphate Synthase (GGPPS) Inhibitors.
J. Med. Chem., 61, 2018
|
|
6C56
 
 | | Crystal structure of mutant human geranylgeranyl pyrophosphate synthase (Y246D) in its Apo form | | Descriptor: | Geranylgeranyl pyrophosphate synthase, {[(2-phenylthieno[2,3-d]pyrimidin-4-yl)amino]methylene}bis(phosphonic acid) | | Authors: | Park, J, Ta, V, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2018-01-15 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Unraveling the Prenylation-Cancer Paradox in Multiple Myeloma with Novel Geranylgeranyl Pyrophosphate Synthase (GGPPS) Inhibitors.
J. Med. Chem., 61, 2018
|
|
6CH4
 
 | |
6CGG
 
 | | Aminoglycoside Phosphotransferase (2'')-Ia in complex with GMPPNP, Magnesium, and Arbekacin | | Descriptor: | Arbekacin, Bifunctional AAC/APH, CHLORIDE ION, ... | | Authors: | Caldwell, S.J, Berghuis, A.M. | | Deposit date: | 2018-02-20 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Plasticity of Aminoglycoside Binding to Antibiotic Kinase APH(2′′)-Ia.
Antimicrob. Agents Chemother., 62, 2018
|
|
5LVC
 
 | | Aichi virus 1: empty particle | | Descriptor: | VP0, VP1, VP3 | | Authors: | Sabin, C, Fuzik, T, Skubnik, K, Palkova, L, Lindberg, A.M, Plevka, P. | | Deposit date: | 2016-09-13 | | Release date: | 2016-12-14 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of Aichi Virus 1 and Its Empty Particle: Clues to Kobuvirus Genome Release Mechanism.
J.Virol., 90, 2016
|
|
5IQG
 
 | | Aminoglycoside Phosphotransferase (2'')-Ia (CTD of AAC(6')-Ie/APH(2'')-Ia) in complex with GDP, Magnesium, and Gentamicin C1 | | Descriptor: | Bifunctional AAC/APH, CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Caldwell, S.J, Berghuis, A.M. | | Deposit date: | 2016-03-10 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Antibiotic Binding Drives Catalytic Activation of Aminoglycoside Kinase APH(2)-Ia.
Structure, 24, 2016
|
|
5IQE
 
 | | Aminoglycoside Phosphotransferase (2'')-Ia (CTD of AAC(6')-Ie/APH(2'')-Ia) in complex with GMPPNP, Magnesium, and Neomycin B | | Descriptor: | Bifunctional AAC/APH, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Caldwell, S.J, Berghuis, A.M. | | Deposit date: | 2016-03-10 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Antibiotic Binding Drives Catalytic Activation of Aminoglycoside Kinase APH(2)-Ia.
Structure, 24, 2016
|
|
1B87
 
 | |
5IQC
 
 | | Aminoglycoside Phosphotransferase (2'')-Ia (CTD of AAC(6')-Ie/APH(2'')-Ia) in complex with GMPPNP, Magnesium, and Gentamicin C1 | | Descriptor: | Bifunctional AAC/APH, CHLORIDE ION, GLYCEROL, ... | | Authors: | Caldwell, S.J, Berghuis, A.M. | | Deposit date: | 2016-03-10 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Antibiotic Binding Drives Catalytic Activation of Aminoglycoside Kinase APH(2)-Ia.
Structure, 24, 2016
|
|
4Y0Y
 
 | | Trypsin in complex with with BPTI | | Descriptor: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | Authors: | Loll, B, Ye, S, Berger, A.A, Muelow, U, Alings, C, Wahl, M.C, Koksch, B. | | Deposit date: | 2015-02-06 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Fluorine teams up with water to restore inhibitor activity to mutant BPTI.
Chem Sci, 6, 2015
|
|
4Y0Z
 
 | | Trypsin in complex with with BPTI mutant AMINOBUTYRIC ACID | | Descriptor: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | Authors: | Loll, B, Ye, S, Berger, A.A, Muelow, U, Alings, C, Wahl, M.C, Koksch, B. | | Deposit date: | 2015-02-06 | | Release date: | 2015-06-24 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Fluorine teams up with water to restore inhibitor activity to mutant BPTI.
Chem Sci, 6, 2015
|
|
4Y11
 
 | | Trypsin in complex with with BPTI mutant (2S)-2-amino-4,4,4-trifluorobutanoic acid | | Descriptor: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | Authors: | Loll, B, Ye, S, Berger, A.A, Muelow, U, Alings, C, Wahl, M.C, Koksch, B. | | Deposit date: | 2015-02-06 | | Release date: | 2015-06-24 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Fluorine teams up with water to restore inhibitor activity to mutant BPTI.
Chem Sci, 6, 2015
|
|
4Y10
 
 | | Trypsin in complex with with BPTI mutant (2S)-2-amino-4,4-difluorobutanoic acid | | Descriptor: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | Authors: | Loll, B, Ye, S, Berger, A.A, Muelow, U, Alings, C, Wahl, M.C, Koksch, B. | | Deposit date: | 2015-02-06 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Fluorine teams up with water to restore inhibitor activity to mutant BPTI.
Chem Sci, 6, 2015
|
|
4YLH
 
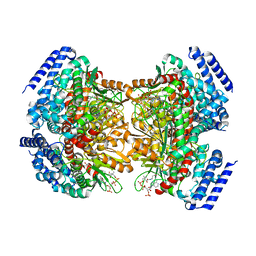 | | Crystal structure of DpgC with bound substrate analog and Xe on oxygen diffusion pathway | | Descriptor: | DpgC, XENON, [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-4-HYDROXY-3-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL (3R)-4-({3-[(2-{[(3,5-DIHYDROXYPHENYL)ACETYL]AMINO}ETHYL)AMINO]-3-OXOPROPYL}AMINO)-3-HYDROXY-2,2-DIMETHYL-4-OXOBUTYL DIHYDROGEN DIPHOSPHATE | | Authors: | Li, K, Di Russo, N.V, Condurso, H.L, Roitberg, A.E, Bruner, S.D. | | Deposit date: | 2015-03-05 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Oxygen diffusion pathways in a cofactor-independent dioxygenase.
Chem Sci, 6, 2015
|
|
8SXN
 
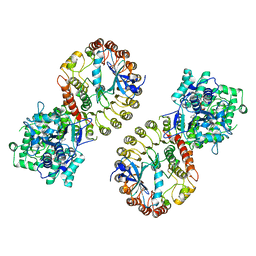 | | Structure of NLRP3 and NEK7 complex | | Descriptor: | 1-[4-(2-oxidanylpropan-2-yl)furan-2-yl]sulfonyl-3-(1,2,3,5-tetrahydro-s-indacen-4-yl)urea, ADENOSINE-5'-TRIPHOSPHATE, NACHT, ... | | Authors: | Yu, X, Matico, R.E, Miller, R, Schoubroeck, B.V, Grauwen, K, Suarez, J, Pietrak, B, Haloi, N, Yin, Y, Tresadern, G.J, Perez-Benito, L, Lindahl, E, Bottelbergs, A, Oehlrich, D, Opdenbosch, N.V, Sharma, S. | | Deposit date: | 2023-05-22 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Cryo-EM structures of NLRP3 reveal its self-activation mechanism
Nat Commun, 2024
|
|
