7MJY
 
 | | MiaB in the complex with s-adenosyl-L-homocysteine and RNA | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FE3-S4 CLUSTER, ... | | Authors: | Esakova, O.A, Grove, T.L, Yennawar, N.H, Arcinas, A.J, Wang, B, Krebs, C, Almo, S.C, Booker, S.J. | | Deposit date: | 2021-04-20 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural basis for tRNA methylthiolation by the radical SAM enzyme MiaB.
Nature, 597, 2021
|
|
7MJZ
 
 | | The structure of MiaB with pentasulfide bridge | | Descriptor: | IRON/SULFUR CLUSTER, PENTASULFIDE-SULFUR, SODIUM ION, ... | | Authors: | Esakova, O.A, Grove, T.L, Yennawar, N.H, Arcinas, A.J, Wang, B, Krebs, C, Almo, S.C, Booker, S.J. | | Deposit date: | 2021-04-20 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural basis for tRNA methylthiolation by the radical SAM enzyme MiaB.
Nature, 597, 2021
|
|
7MJW
 
 | | Methylated MiaB in the complex with 5'-deoxyadenosine, methionine and RNA | | Descriptor: | 5'-DEOXYADENOSINE, FE3-S4 methylated cluster, IRON/SULFUR CLUSTER, ... | | Authors: | Esakova, O.A, Grove, T.L, Yennawar, N.H, Arcinas, A.J, Wang, B, Krebs, C, Almo, S.C, Booker, S.J. | | Deposit date: | 2021-04-20 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for tRNA methylthiolation by the radical SAM enzyme MiaB.
Nature, 597, 2021
|
|
7MJX
 
 | | MiaB in the complex with 5'-deoxyadenosine, methionine and RNA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-DEOXYADENOSINE, FE3-S4 CLUSTER, ... | | Authors: | Esakova, O.A, Grove, T.L, Yennawar, N.H, Arcinas, A.J, Wang, B, Krebs, C, Almo, S.C, Booker, S.J. | | Deposit date: | 2021-04-20 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for tRNA methylthiolation by the radical SAM enzyme MiaB.
Nature, 597, 2021
|
|
5IIH
 
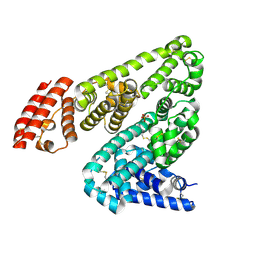 | | Crystal structure of Equine Serum Albumin in the presence of 2.5 mM zinc at pH 7.4 | | Descriptor: | SULFATE ION, Serum albumin, ZINC ION | | Authors: | Handing, K.B, Shabalin, I.G, Cooper, D.R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2016-03-01 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Circulatory zinc transport is controlled by distinct interdomain sites on mammalian albumins.
Chem Sci, 7, 2016
|
|
7N7I
 
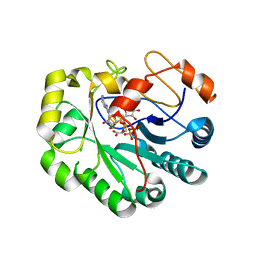 | | X-ray crystal structure of Viperin-like enzyme from Trichoderma virens | | Descriptor: | IRON/SULFUR CLUSTER, S-ADENOSYLMETHIONINE, URIDINE 5'-TRIPHOSPHATE, ... | | Authors: | Grove, T.L, Almo, S.C, Bonanno, J.B, Lachowicz, J.C, Gizzi, A.G. | | Deposit date: | 2021-06-10 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structural Insight into the Substrate Scope of Viperin and Viperin-like Enzymes from Three Domains of Life.
Biochemistry, 60, 2021
|
|
7N7H
 
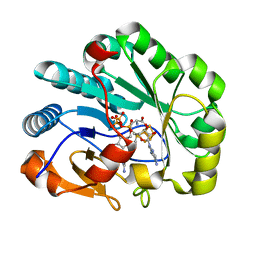 | | X-ray crystal structure of Viperin-like enzyme from Nematostella vectensis | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, IRON/SULFUR CLUSTER, S-ADENOSYLMETHIONINE, ... | | Authors: | Grove, T.L, Almo, S.C, Bonanno, J.B, Lachowicz, J.C, Gizzi, A.G. | | Deposit date: | 2021-06-10 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural Insight into the Substrate Scope of Viperin and Viperin-like Enzymes from Three Domains of Life.
Biochemistry, 60, 2021
|
|
7N2W
 
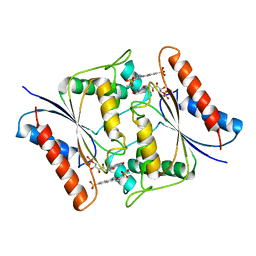 | | The crystal structure of an FMN-dependent NADH-azoreductase, AzoA in complex with Red 40 | | Descriptor: | 6-hydroxy-5-[(E)-(2-methoxy-5-methyl-4-sulfophenyl)diazenyl]naphthalene-2-sulfonic acid, FLAVIN MONONUCLEOTIDE, FMN dependent NADH:quinone oxidoreductase | | Authors: | Arcinas, A.J, Fedorov, E, Kelly, L, Almo, S.C, Ghosh, A. | | Deposit date: | 2021-05-30 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Uncovering a novel mechanism of enzyme activation in multimeric azoreductases
To Be Published
|
|
7N2X
 
 | | The crystal structure of an FMN-dependent NADH:quinone oxidoreductase, AzoR from Escherichia coli | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, 2-AMINO-ACRYLIC ACID, ... | | Authors: | Arcinas, A.J, Fedorov, E, Kelly, L, Almo, S.C, Ghosh, A. | | Deposit date: | 2021-05-30 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Uncovering a novel mechanism of enzyme activation in multimeric azoreductases
To Be Published
|
|
5BP7
 
 | | Crystal structure of SAM-dependent methyltransferase from Geobacter sulfurreducens in complex with S-Adenosyl-L-homocysteine | | Descriptor: | CHLORIDE ION, S-ADENOSYL-L-HOMOCYSTEINE, SAM-dependent methyltransferase | | Authors: | Kutner, J, Shabalin, I.G, Mason, D.V, Handing, K.B, Gasiorowska, O.A, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-10 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of SAM-dependent methyltransferase from Geobacter sulfurreducens in complex with S-Adenosyl-L-homocysteine
to be published
|
|
5BR1
 
 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM AGROBACTERIUM VITIS S4 (Avi_5305, TARGET EFI-511224) WITH BOUND ALPHA-D-GALACTOSAMINE | | Descriptor: | 2-amino-2-deoxy-alpha-D-galactopyranose, ABC transporter, binding protein | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-05-29 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of an ABC transporter solute-binding protein specific for the amino sugars glucosamine and galactosamine.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5BP9
 
 | | Crystal structure of SAM-dependent methyltransferase from Bacteroides fragilis in complex with S-Adenosyl-L-homocysteine | | Descriptor: | 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Putative methyltransferase protein, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Gasiorowska, O.A, Shabalin, I.G, Handing, K.B, Cymborowski, M.T, Mason, D.V, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-10 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of SAM-dependent methyltransferase fromBacteroides fragilis in complex with S-Adenosyl-L-homocysteine
to be published
|
|
5C7H
 
 | | Crystal structure of aldo-keto reductase from Sinorhizobium meliloti 1021 in complex with NADPH | | Descriptor: | Aldo-keto reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Gasiorowska, O.A, Shabalin, I.G, Handing, K.B, Seidel, R, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-06-24 | | Release date: | 2015-07-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of aldo-keto reductase from Sinorhizobium meliloti 1021 in complex with NADPH
to be published
|
|
5CI5
 
 | | Crystal Structure of an ABC transporter Solute Binding Protein from Thermotoga Lettingae TMO (Tlet_1705, TARGET EFI-510544) bound with alpha-D-Tagatose | | Descriptor: | 1,2-ETHANEDIOL, Extracellular solute-binding protein family 1, PENTAETHYLENE GLYCOL, ... | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-07-11 | | Release date: | 2015-07-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal Structure of an ABC transporter Solute Binding Protein from Thermotoga Lettingae TMO (Tlet_1705, TARGET EFI-510544) bound with alpha-D-Tagatose
To be published
|
|
5BXY
 
 | | Crystal structure of RNA methyltransferase from Salinibacter ruber in complex with S-Adenosyl-L-homocysteine | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, RNA methyltransferase, ... | | Authors: | Handing, K.B, LaRowe, C, Shabalin, I.G, Stead, M, Hillerich, B.S, Ahmed, M, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-06-09 | | Release date: | 2015-07-01 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of RNA methylase family protein from Salinibacterruber in complex with S-Adenosyl-L-homocysteine.
to be published
|
|
5C9G
 
 | | Crystal Structure of a Putative enoyl-CoA hydratase/isomerase family protein from Hyphomonas neptunium | | Descriptor: | D-MALATE, Enoyl-CoA hydratase/isomerase family protein, TETRAETHYLENE GLYCOL | | Authors: | Szlachta, K, Cooper, D.R, Chapman, H.C, Cymbrowski, M.T, Stead, M, Hillerich, B.S, Ahmed, M, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, Hammonds, J, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-06-26 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of a Putative enoyl-CoA hydratase/isomerase family protein from Hyphomonas neptunium
to be published
|
|
5CGS
 
 | |
5CHM
 
 | | CRYSTAL STRUCTURE OF Fox-4 cephamycinase complexed with ceftazidime BATSI (LP06) | | Descriptor: | ACETATE ION, Beta-lactamase, PINACOL[[2-AMINO-ALPHA-(1-CARBOXY-1-METHYLETHOXYIMINO)-4-THIAZOLEACETYL]AMINO]METHANEBORONATE, ... | | Authors: | Malashkevich, V.N, Toro, R, Lefurgy, S, Almo, S.C. | | Deposit date: | 2015-07-10 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of FOX-4 Cephamycinase in Complex with Transition-State Analog Inhibitors.
Biomolecules, 10, 2020
|
|
5CHU
 
 | | CRYSTAL STRUCTURE OF Fox-4 cephamycinase complexed with sulfate | | Descriptor: | ACETATE ION, Beta-lactamase, SULFATE ION, ... | | Authors: | Malashkevich, V.N, Toro, R, Lefurgy, S, Almo, S.C. | | Deposit date: | 2015-07-10 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | CRYSTAL STRUCTURE OF Fox-4 cephamycinase complexed with sulfate
To Be Published
|
|
5CGW
 
 | | CRYSTAL STRUCTURE OF Fox-4 cephamycinase mutant Y150F | | Descriptor: | ACETATE ION, Beta-lactamase, ZINC ION | | Authors: | Malashkevich, V.N, Toro, R, Lefurgy, S, Almo, S.C. | | Deposit date: | 2015-07-09 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | FOX-4 cephamycinase: an analysis of structure and function.
Antimicrob.Agents Chemother., 2015
|
|
5CHJ
 
 | | CRYSTAL STRUCTURE OF Fox-4 cephamycinase complexed with cephalothin BATSI (SM23) | | Descriptor: | (1R)-1-(2-THIENYLACETYLAMINO)-1-(3-CARBOXYPHENYL)METHYLBORONIC ACID, ACETATE ION, Beta-lactamase, ... | | Authors: | Malashkevich, V.N, Toro, R, Lefurgy, S, Almo, S.C. | | Deposit date: | 2015-07-10 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.358 Å) | | Cite: | Structures of FOX-4 Cephamycinase in Complex with Transition-State Analog Inhibitors.
Biomolecules, 10, 2020
|
|
5CGX
 
 | | CRYSTAL STRUCTURE OF Fox-4 cephamycinase mutant Y150F complexed with cefoxitin | | Descriptor: | (2R)-2-{(1S)-1-methoxy-2-oxo-1-[(thiophen-2-ylacetyl)amino]ethyl}-5-methylidene-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase, SODIUM ION, ... | | Authors: | Malashkevich, V.N, Toro, R, Lefurgy, S, Almo, S.C. | | Deposit date: | 2015-07-09 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | FOX-4 cephamycinase: an analysis of structure and function.
Antimicrob.Agents Chemother., 2015
|
|
5CM6
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM PSEUDOALTEROMONAS ATLANTICA T6c(Patl_2292, TARGET EFI-510180) WITH BOUND SODIUM AND PYRUVATE | | Descriptor: | PYRUVIC ACID, SODIUM ION, TRAP dicarboxylate transporter-DctP subunit | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-07-16 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM PSEUDOALTEROMONAS ATLANTICA T6c(Patl_2292, TARGET EFI-510180) WITH BOUND SODIUM AND PYRUVATE
To be published
|
|
4X24
 
 | | Crystal structure of Vibrio cholerae 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with methylthio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)METHYL]-4-[(METHYLSULFANYL)METHYL]PYRROLIDIN-3-OL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, TRIETHYLENE GLYCOL | | Authors: | Cameron, S.A, Thomas, K, Almo, S.C, Schramm, V.L. | | Deposit date: | 2014-11-25 | | Release date: | 2015-08-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Active site and remote contributions to catalysis in methylthioadenosine nucleosidases.
Biochemistry, 54, 2015
|
|
5DMH
 
 | | Crystal structure of a domain of unknown function (DUF1537) from Ralstonia eutropha H16 (H16_A1561), Target EFI-511666, complex with ADP. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Uncharacterized protein conserved in bacteria | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-09-08 | | Release date: | 2015-10-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a domain of unknown function (DUF1537) from Ralstonia eutropha H16 (H16_A1561), Target EFI-511666, complex with ADP.
To be published
|
|
