5UYQ
 
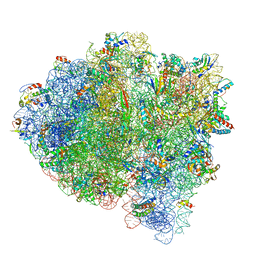 | | 70S ribosome bound with near-cognate ternary complex base-paired to A site codon, closed 30S (Structure III-nc) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2017-02-24 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Ensemble cryo-EM elucidates the mechanism of translation fidelity
Nature, 546, 2017
|
|
4AWD
 
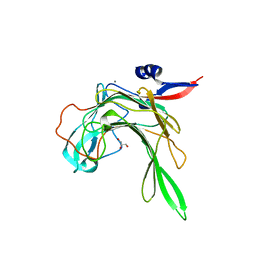 | | Crystal structure of the beta-porphyranase BpGH16B (BACPLE_01689) from the human gut bacterium Bacteroides plebeius | | Descriptor: | BETA-PORPHYRANASE, CALCIUM ION, GLYCEROL | | Authors: | Hehemann, J.H, Kelly, A.G, Pudlo, N.A, Martens, E.C, Boraston, A.B. | | Deposit date: | 2012-06-01 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bacteria of the human gut microbiome catabolize red seaweed glycans with carbohydrate-active enzyme updates from extrinsic microbes.
Proc. Natl. Acad. Sci. U.S.A., 109, 2012
|
|
5UPI
 
 | |
5V1W
 
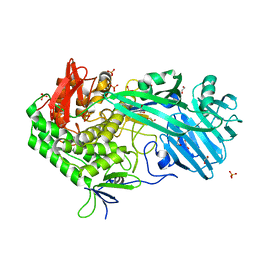 | | Crystal structure of BhGH81 in complex with laminaro-biose | | Descriptor: | 1,2-ETHANEDIOL, Glycoside Hydrolase, PHOSPHATE ION, ... | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2017-03-02 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The quaternary structure of beta-1,3-glucan contributes to its recognition and hydrolysis by a multimodular family 81 glycoside hydrolase
Structure, 2017
|
|
5UWN
 
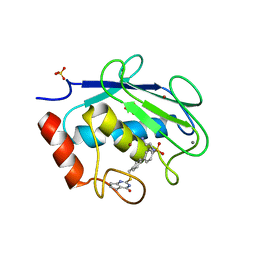 | | Matrix metalloproteinase-13 complexed with selective inhibitor compound 10d | | Descriptor: | CALCIUM ION, Collagenase 3, N-(2-aminoethyl)-4'-(((4-oxo-4,5,6,7-tetrahydro-3H-cyclopenta[d]pyrimidin-2-yl)thio)methyl)-[1,1'-biphenyl]-4-sulfonami de, ... | | Authors: | Taylor, A.B, Cao, X, Hart, P.J. | | Deposit date: | 2017-02-21 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-Based Design and Synthesis of Potent and Selective Matrix Metalloproteinase 13 Inhibitors.
J. Med. Chem., 60, 2017
|
|
5UYL
 
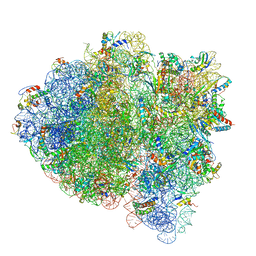 | | 70S ribosome bound with cognate ternary complex base-paired to A site codon (Structure II) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2017-02-24 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Ensemble cryo-EM elucidates the mechanism of translation fidelity.
Nature, 546, 2017
|
|
5UPM
 
 | |
5VYH
 
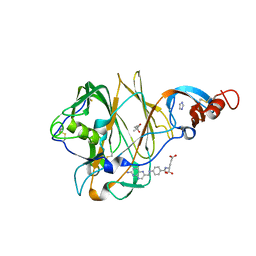 | | Crystal Structure of MERS-CoV S1 N-terminal Domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, FOLIC ACID, ... | | Authors: | Wang, N, Wrapp, D, Pallesen, J, Ward, A.B, McLellan, J.S. | | Deposit date: | 2017-05-25 | | Release date: | 2017-08-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4AK5
 
 | | Native crystal structure of BpGH117 | | Descriptor: | 1,2-ETHANEDIOL, ANHYDRO-ALPHA-L-GALACTOSIDASE, CHLORIDE ION, ... | | Authors: | Hehemann, J.H, Smyth, L, Yadav, A, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2012-02-21 | | Release date: | 2012-03-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Analysis of Keystone Enzyme in Agar Hydrolysis Provides Insight Into the Degradation (of a Polysaccharide from) Red Seaweeds.
J.Biol.Chem., 287, 2012
|
|
4B28
 
 | | Crystal structure of DMSP lyase RdDddP from Roseobacter denitrificans | | Descriptor: | FE (III) ION, METALLOPEPTIDASE, FAMILY M24, ... | | Authors: | Hehemann, J.H, Law, A, Redecke, L, Boraston, A.B. | | Deposit date: | 2012-07-12 | | Release date: | 2012-07-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Structure of Rddddp from Roseobacter Denitrificans Reveals that Dmsp Lyases in the Dddp-Family are Metalloenzymes.
Plos One, 9, 2014
|
|
4AZ6
 
 | | Differential inhibition of the tandem GH20 catalytic modules in the pneumococcal exo-beta-D-N-acetylglucosaminidase, StrH | | Descriptor: | 1,2-ETHANEDIOL, 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, BETA-N-ACETYLHEXOSAMINIDASE, ... | | Authors: | Pluvinage, B, Stubbs, K.A, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2012-06-23 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Inhibition of the Family 20 Glycoside Hydrolase Catalytic Modules in the Streptococcus Pneumoniae Exo-Beta-D-N-Acetylglucosaminidase, Strh.
Org.Biomol.Chem., 11, 2013
|
|
4AW7
 
 | | BpGH86A: A beta-porphyranase of glycoside hydrolase family 86 from the human gut bacterium Bacteroides plebeius | | Descriptor: | 1,2-ETHANEDIOL, 6-O-sulfo-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-6-O-sulfo-alpha-L-galactopyranose-(1-3)-alpha-D-galactopyranose, CALCIUM ION, ... | | Authors: | Hehemann, J.H, Kelly, A.G, Pudlo, N.A, Martens, E.C, Boraston, A.B. | | Deposit date: | 2012-06-01 | | Release date: | 2012-07-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Bacteria of the human gut microbiome catabolize red seaweed glycans with carbohydrate-active enzyme updates from extrinsic microbes.
Proc. Natl. Acad. Sci. U.S.A., 109, 2012
|
|
5VN8
 
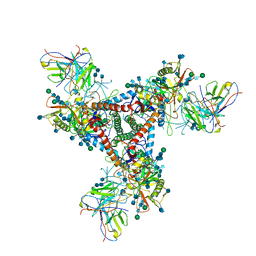 | | Cryo-EM model of B41 SOSIP.664 in complex with fragment antigen binding variable domain of b12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Ozorowski, G, Pallesen, J, Ward, A.B, Cottrell, C.A. | | Deposit date: | 2017-04-28 | | Release date: | 2017-07-12 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Open and closed structures reveal allostery and pliability in the HIV-1 envelope spike.
Nature, 547, 2017
|
|
5VQZ
 
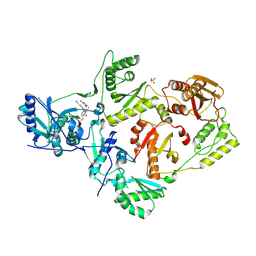 | | Crystal Structure of HIV-1 Reverse Transcriptase (K103N, Y181C) Variant in Complex with 2-chloro-N-(6-cyano-3-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-4-methylnaphthalen-1-yl)-N-methylacetamide (JLJ686), a Non-nucleoside Inhibitor | | Descriptor: | N-(6-cyano-3-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-4-methylnaphthalen-1-yl)-N-methylacetamide, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Buckingham, A.B, Chan, A.H, Anderson, K.S. | | Deposit date: | 2017-05-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Covalent inhibitors for eradication of drug-resistant HIV-1 reverse transcriptase: From design to protein crystallography.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4AAX
 
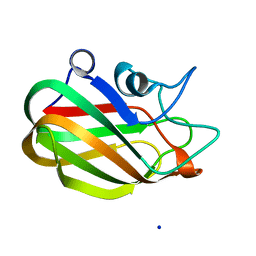 | | CpGH89CBM32-5, from Clostridium perfringens, in complex with N- acetylgalactosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, ALPHA-N-ACETYLGLUCOSAMINIDASE, CALCIUM ION, ... | | Authors: | Ficko-Blean, E, Stuart, C.P, Suits, M.D, Cid, M, Tessier, M, Woods, R.J, Boraston, A.B. | | Deposit date: | 2011-12-05 | | Release date: | 2012-04-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Carbohydrate Recognition by an Architecturally Complex Alpha-N-Acetylglucosaminidase from Clostridium Perfringens.
Plos One, 7, 2012
|
|
4A34
 
 | |
4A42
 
 | | CpGH89CBM32-6 produced by Clostridium perfringens | | Descriptor: | ALPHA-N-ACETYLGLUCOSAMINIDASE FAMILY PROTEIN, CALCIUM ION | | Authors: | Ficko-Blean, E, Stuart, C.P, Suits, M.D, Cid, M, Tessier, M, Woods, R.J, Boraston, A.B. | | Deposit date: | 2011-10-06 | | Release date: | 2012-04-04 | | Last modified: | 2012-04-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Carbohydrate Recognition by an Architecturally Complex Alpha-N-Acetylglucosaminidase from Clostridium Perfringens.
Plos One, 7, 2012
|
|
5UPO
 
 | |
4BFQ
 
 | | Assembly of a triple pi-stack of ligands in the binding site of Aplysia californica acetylcholine binding protein (AChBP) | | Descriptor: | 4,6-dimethyl-N'-(3-pyridin-2-ylisoquinolin-1-yl)pyrimidine-2-carboximidamide, GLYCEROL, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Stornaiuolo, M, De Kloe, G.E, Rucktooa, P, Fish, A, van Elk, R, Edink, E.S, Bertrand, D, Smit, A.B, de Esch, I.J.P, Sixma, T.K. | | Deposit date: | 2013-03-21 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Assembly of a Pi-Pi Stack of Ligands in the Binding Site of an Acetylcholine Binding Protein
Nat.Commun., 4, 2013
|
|
4D6G
 
 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98) in complex with the blood group A-trisaccharide (L19 mutant) | | Descriptor: | 1,2-ETHANEDIOL, GLYCOSIDE HYDROLASE, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]alpha-D-galactopyranose | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-11 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
5UWL
 
 | | Matrix metalloproteinase-13 complexed with selective inhibitor compound (S)-17a | | Descriptor: | (S)-N-(3-methyl-1-(methylamino)-1-oxobutan-2-yl)-5-(4-(((4-oxo-4,5,6,7-tetrahydro-3H-cyclopenta[d]pyrimidin-2-yl)thio)methyl)phenyl)furan-2-carboxamide, CALCIUM ION, Collagenase 3, ... | | Authors: | Taylor, A.B, Cao, X, Hart, P.J. | | Deposit date: | 2017-02-21 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure-Based Design and Synthesis of Potent and Selective Matrix Metalloproteinase 13 Inhibitors.
J. Med. Chem., 60, 2017
|
|
4D71
 
 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98) in complex with the type 2 blood group A-tetrasaccharide (E558A X02 mutant) | | Descriptor: | 1,2-ETHANEDIOL, GLYCOSIDE HYDROLASE, SULFATE ION, ... | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-19 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
5TA0
 
 | | Crystal structure of BuGH86E322Q in complex with neoagarooctaose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Pluvinage, B, Boraston, A.B, Abbott, W.D. | | Deposit date: | 2016-09-09 | | Release date: | 2017-09-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular basis of an agarose metabolic pathway acquired by a human intestinal symbiont.
Nat Commun, 9, 2018
|
|
5T9A
 
 | | Crystal structure of BuGH2Cwt | | Descriptor: | 1,2-ETHANEDIOL, Glycoside Hydrolase, SULFATE ION | | Authors: | Pluvinage, B, Boraston, A.B, Abbott, W.D. | | Deposit date: | 2016-09-09 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis of an agarose metabolic pathway acquired by a human intestinal symbiont.
Nat Commun, 9, 2018
|
|
5T7A
 
 | | Crystal structure of Br derivative BhCBM56 | | Descriptor: | 1,2-ETHANEDIOL, BH0236 protein, BROMIDE ION | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2016-09-02 | | Release date: | 2017-08-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Properties of a family 56 carbohydrate-binding module and its role in the recognition and hydrolysis of beta-1,3-glucan.
J. Biol. Chem., 292, 2017
|
|
