4UAP
 
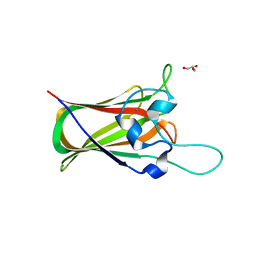 | | X-ray structure of GH31 CBM32-2 bound to GalNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Grondin, J.M, Abe, K, Boraston, A.B, Smith, S.P. | | Deposit date: | 2014-08-11 | | Release date: | 2015-10-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Diverse modes of galacto-specific carbohydrate recognition by a family 31 glycoside hydrolase from Clostridium perfringens.
PLoS ONE, 12, 2017
|
|
4UTF
 
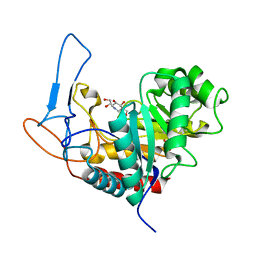 | | Structure of the GH99 endo-alpha-mannosidase from Bacteroides xylanisolvens in complex with mannose-alpha-1,3-isofagomine and alpha- 1,2-mannobiose | | Descriptor: | 1,2-ETHANEDIOL, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, GLYCOSYL HYDROLASE FAMILY 71, ... | | Authors: | Cuskin, F, Lowe, E.C, Temple, M.J, Zhu, Y, Pudlo, N.A, Cameron, E.A, Urs, K, Thompson, A.J, Cartmell, A, Rogowski, A, Tolbert, T, Piens, K, Bracke, D, Vervecken, W, Hakki, Z, Speciale, G, Munoz-Munoz, J.L, Pena, M.J, McLean, R, Suits, M.D, Boraston, A.B, Atherly, T, Ziemer, C.J, Williams, S.J, Davies, G.J, Abbott, D.W, Martens, E.C, Gilbert, H.J. | | Deposit date: | 2014-07-21 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Human Gut Bacteroidetes Can Utilize Yeast Mannan Through a Selfish Mechanism.
Nature, 517, 2015
|
|
5FGJ
 
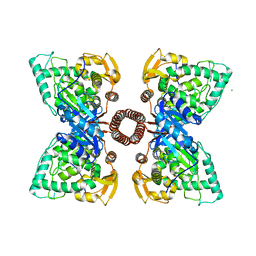 | | Structure of tetrameric rat phenylalanine hydroxylase, residues 1-453 | | Descriptor: | FE (III) ION, MAGNESIUM ION, Phenylalanine-4-hydroxylase | | Authors: | Taylor, A.B, Roberts, K.M, Fitzpatrick, P.F. | | Deposit date: | 2015-12-20 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Domain Movements upon Activation of Phenylalanine Hydroxylase Characterized by Crystallography and Chromatography-Coupled Small-Angle X-ray Scattering.
J.Am.Chem.Soc., 138, 2016
|
|
5FR0
 
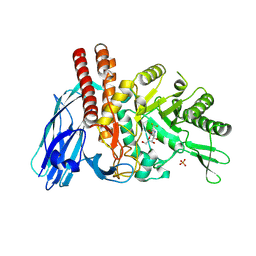 | | The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens | | Descriptor: | 2-deoxy-2-[(difluoroacetyl)amino]-beta-D-galactopyranose, BETA-N-ACETYLGALACTOSAMINIDASE, PHOSPHATE ION | | Authors: | Noach, I, Pluvinage, B, Laurie, C, Abe, K.T, Alteen, M, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-12-14 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Details of Glycolipid Glycan Hydrolysis by the Structural Analysis of a Family 123 Glycoside Hydrolase from Clostridium Perfringens
J.Mol.Biol., 428, 2016
|
|
5FQG
 
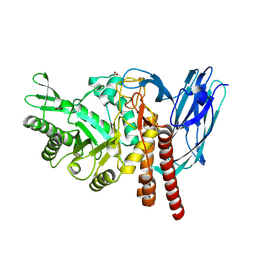 | | The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-beta-D-galactopyranose, BETA-N-ACETYLGALACTOSAMINIDASE, FORMIC ACID | | Authors: | Noach, I, Pluvinage, B, Laurie, C, Abe, K.T, Alteen, M, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-12-10 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Details of Glycolipid Glycan Hydrolysis by the Structural Analysis of a Family 123 Glycoside Hydrolase from Clostridium Perfringens
J.Mol.Biol., 428, 2016
|
|
5FUU
 
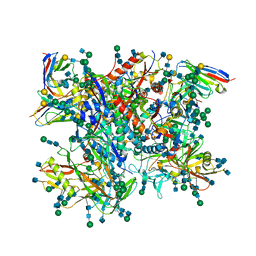 | | Ectodomain of cleaved wild type JR-FL EnvdCT trimer in complex with PGT151 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lee, J.H, Ward, A.B. | | Deposit date: | 2016-01-29 | | Release date: | 2016-03-09 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-Em Structure of a Native, Fully Glycosylated and Cleaved HIV-1 Envelope Trimer
Science, 351, 2016
|
|
1BIZ
 
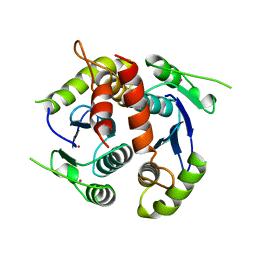 | | HIV-1 INTEGRASE CORE DOMAIN | | Descriptor: | CACODYLATE ION, HIV-1 INTEGRASE | | Authors: | Goldgur, Y, Dyda, F, Hickman, A.B, Jenkins, T.M, Craigie, R, Davies, D.R. | | Deposit date: | 1998-06-21 | | Release date: | 1998-08-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
5FNA
 
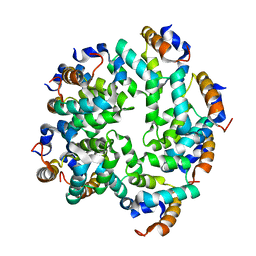 | | Cryo-EM reconstruction of caspase-1 CARD | | Descriptor: | Caspase-1 | | Authors: | Li, Y, Lu, A, Schmidt, F.I, Yin, Q, Chen, S, Fu, T.M, Tong, A.B, Ploegh, H.L, Mao, Y, Wu, H. | | Deposit date: | 2015-11-11 | | Release date: | 2016-03-30 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Molecular Basis of Caspase-1 Polymerization and its Inhibition by a Novel Capping Mechanism
Nat.Struct.Mol.Biol., 23, 2016
|
|
5FQF
 
 | | The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, BETA-N-ACETYLGALACTOSAMINIDASE, FORMIC ACID | | Authors: | Noach, I, Pluvinage, B, Laurie, C, Abe, K.T, Alteen, M, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-12-10 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Details of Glycolipid Glycan Hydrolysis by the Structural Analysis of a Family 123 Glycoside Hydrolase from Clostridium Perfringens
J.Mol.Biol., 428, 2016
|
|
5A6A
 
 | | GH20C, Beta-hexosaminidase from Streptococcus pneumoniae in complex with NGT | | Descriptor: | 1,2-ETHANEDIOL, 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, N-ACETYL-BETA-D-GLUCOSAMINIDASE | | Authors: | Cid, M, Robb, C.S, Higgins, M.A, Boraston, A.B. | | Deposit date: | 2015-06-24 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | A Second beta-Hexosaminidase Encoded in the Streptococcus pneumoniae Genome Provides an Expanded Biochemical Ability to Degrade Host Glycans.
J. Biol. Chem., 290, 2015
|
|
5A57
 
 | | The structure of GH101 from Streptococcus pneumoniae TIGR4 in complex with PUGT | | Descriptor: | (Z)-[(3R,4R,5R,6R)-3-acetamido-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-2-ylidene]amino] N-phenylcarbamate, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Gregg, K.J, Suits, M.D.L, Deng, L, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-06-16 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural Analysis of a Family 101 Glycoside Hydrolase in Complex with Carbohydrates Reveals Insights Into its Mechanism.
J.Biol.Chem., 290, 2015
|
|
1EHN
 
 | | CRYSTAL STRUCTURE OF CHITINASE A MUTANT E315Q COMPLEXED WITH OCTA-N-ACETYLCHITOOCTAOSE (NAG)8. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE A | | Authors: | Papanikolau, Y, Prag, G, Tavlas, G, Vorgias, C.E, Oppenheim, A.B, Petratos, K. | | Deposit date: | 2000-02-22 | | Release date: | 2001-02-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High resolution structural analyses of mutant chitinase A complexes with substrates provide new insight into the mechanism of catalysis.
Biochemistry, 40, 2001
|
|
1ETZ
 
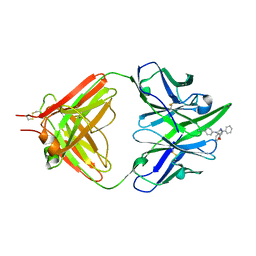 | | THE THREE-DIMENSIONAL STRUCTURE OF AN ANTI-SWEETENER FAB, NC10.14, SHOWS THE EXTENT OF STRUCTURAL DIVERSITY IN ANTIGEN RECOGNITION BY IMMUNOGLOBULINS | | Descriptor: | FAB NC10.14 - HEAVY CHAIN, FAB NC10.14 - LIGHT CHAIN, N-(P-CYANOPHENYL)-N'-DIPHENYLMETHYL-GUANIDINE-ACETIC ACID | | Authors: | Guddat, L.W, Shan, L, Broomell, C, Ramsland, P.A, Fan, Z, Anchin, J.M, Linthicum, D.S, Edmundson, A.B. | | Deposit date: | 2000-04-13 | | Release date: | 2000-10-18 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The three-dimensional structure of a complex of a murine Fab (NC10. 14) with a potent sweetener (NC174): an illustration of structural diversity in antigen recognition by immunoglobulins.
J.Mol.Biol., 302, 2000
|
|
5I08
 
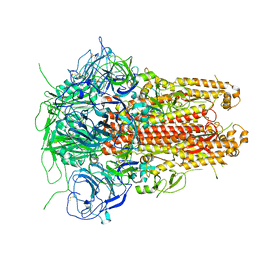 | | Prefusion structure of a human coronavirus spike protein | | Descriptor: | Spike glycoprotein,Foldon chimera | | Authors: | Kirchdoerfer, R.N, Cottrell, C.A, Wang, N, Pallesen, J, Yassine, H.M, Turner, H.L, Corbett, K.S, Graham, B.S, McLellan, J.S, Ward, A.B. | | Deposit date: | 2016-02-03 | | Release date: | 2016-03-02 | | Last modified: | 2020-04-22 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Pre-fusion structure of a human coronavirus spike protein.
Nature, 531, 2016
|
|
5A56
 
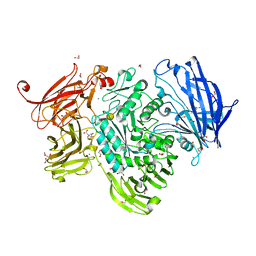 | | The structure of GH101 from Streptococcus pneumoniae TIGR4 in complex with 1-O-methyl-T-antigen | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CITRIC ACID, ... | | Authors: | Gregg, K.J, Suits, M.D.L, Deng, L, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-06-16 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Analysis of a Family 101 Glycoside Hydrolase in Complex with Carbohydrates Reveals Insights into Its Mechanism.
J.Biol.Chem., 290, 2015
|
|
5FQ0
 
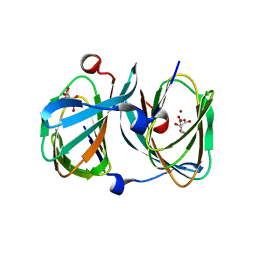 | | The structure of KdgF from Halomonas sp. | | Descriptor: | CITRATE ANION, KDGF, NICKEL (II) ION, ... | | Authors: | Hobbs, J.K, Lee, S.M, Robb, M, Hof, F, Barr, C, Abe, K.T, Hehemann, J.H, McLean, R, Abbott, D.W, Boraston, A.B. | | Deposit date: | 2015-12-03 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kdgf, the Missing Link in the Microbial Metabolism of Uronate Sugars from Pectin and Alginate.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1BIU
 
 | | HIV-1 INTEGRASE CORE DOMAIN COMPLEXED WITH MG++ | | Descriptor: | HIV-1 INTEGRASE, MAGNESIUM ION | | Authors: | Goldgur, Y, Dyda, F, Hickman, A.B, Jenkins, T.M, Craigie, R, Davies, D.R. | | Deposit date: | 1998-06-19 | | Release date: | 1998-08-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
4WYM
 
 | | Structural basis of HIV-1 capsid recognition by CPSF6 | | Descriptor: | Capsid protein p24, ISOFORM 2 OF CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR SUBUNIT 6 | | Authors: | Battacharya, A, Taylor, A.B, Hart, P.J, Ivanov, D.N. | | Deposit date: | 2014-11-17 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of HIV-1 capsid recognition by PF74 and CPSF6.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5AC4
 
 | | GH20C, Beta-hexosaminidase from Streptococcus pneumoniae in complex with GalNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, N-ACETYL-BETA-D-GLUCOSAMINIDASE | | Authors: | Cid, M, Robb, C.S, Higgins, M.A, Boraston, A.B. | | Deposit date: | 2015-08-11 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | A Second beta-Hexosaminidase Encoded in the Streptococcus pneumoniae Genome Provides an Expanded Biochemical Ability to Degrade Host Glycans.
J. Biol. Chem., 290, 2015
|
|
5A69
 
 | | GH20C, Beta-hexosaminidase from Streptococcus pneumoniae in complex with Gal-PUGNAc | | Descriptor: | N-ACETYL-BETA-D-GLUCOSAMINIDASE, [(Z)-[(3R,4R,5R,6R)-3-acetamido-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-2-ylidene]amino] N-phenylcarbamate | | Authors: | Cid, M, Robb, C.S, Higgins, M.A, Boraston, A.B. | | Deposit date: | 2015-06-24 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Second beta-Hexosaminidase Encoded in the Streptococcus pneumoniae Genome Provides an Expanded Biochemical Ability to Degrade Host Glycans.
J. Biol. Chem., 290, 2015
|
|
5A58
 
 | | The structure of GH101 D764N mutant from Streptococcus pneumoniae TIGR4 in complex with serinyl T-antigen | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CITRIC ACID, ... | | Authors: | Gregg, K.J, Suits, M.D.L, Deng, L, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-06-16 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Analysis of a Family 101 Glycoside Hydrolase in Complex with Carbohydrates Reveals Insights Into its Mechanism.
J.Biol.Chem., 290, 2015
|
|
5A6B
 
 | | GH20C, Beta-hexosaminidase from Streptococcus pneumoniae in complex with PUGNAc | | Descriptor: | MAGNESIUM ION, N-ACETYL-BETA-D-GLUCOSAMINIDASE, O-(2-ACETAMIDO-2-DEOXY D-GLUCOPYRANOSYLIDENE) AMINO-N-PHENYLCARBAMATE | | Authors: | Cid, M, Robb, C.S, Higgins, M.A, Boraston, A.B. | | Deposit date: | 2015-06-24 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A Second beta-Hexosaminidase Encoded in the Streptococcus pneumoniae Genome Provides an Expanded Biochemical Ability to Degrade Host Glycans.
J. Biol. Chem., 290, 2015
|
|
5A6J
 
 | | GH20C, Beta-hexosaminidase from Streptococcus pneumoniae | | Descriptor: | 1,2-ETHANEDIOL, N-ACETYL-BETA-D-GLUCOSAMINIDASE | | Authors: | Cid, M, Robb, C.S, Higgins, M.A, Boraston, A.B. | | Deposit date: | 2015-06-26 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A Second beta-Hexosaminidase Encoded in the Streptococcus pneumoniae Genome Provides an Expanded Biochemical Ability to Degrade Host Glycans.
J. Biol. Chem., 290, 2015
|
|
5A6K
 
 | | GH20C, Beta-hexosaminidase from Streptococcus pneumoniae in complex with Gal-NGT | | Descriptor: | (2S,3aR,5R,6R,7R,7aR)-5-(hydroxymethyl)-2-methyl-2,3a,5,6,7,7a-hexahydro-1H-pyrano[3,2-d][1,3]thiazole-6,7-diol, GH20C | | Authors: | Cid, M, Robb, C.S, Higgins, M.A, Boraston, A.B. | | Deposit date: | 2015-06-26 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Second Beta-Hexosaminidase Encoded in the Streptococcus Pneumoniae Genome Provides an Expanded Biochemical Ability to Degrade Host Glycans
J.Biol.Chem., 290, 2015
|
|
5AN1
 
 | |
