8I83
 
 | |
8IKU
 
 | |
5WCI
 
 | | Human MYST histone acetyltransferase 1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Histone acetyltransferase KAT8, ... | | Authors: | Dong, A, Zeng, H, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Zheng, Y.G, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-30 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Human MYST histone acetyltransferase 1
to be published
|
|
8J0I
 
 | | Aldo-keto reductase KmAKR | | Descriptor: | NADPH-dependent alpha-keto amide reductase, SODIUM ION | | Authors: | Xu, S.Y, Zhou, L, Xu, Y, Wang, Y.J, Zheng, Y.G. | | Deposit date: | 2023-04-11 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Aldo-keto reductase KmAKR from Kluyveromyces marxianus
To Be Published
|
|
4X41
 
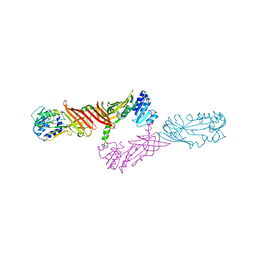 | | Crystal Structure of Protein Arginine Methyltransferase PRMT8 | | Descriptor: | Protein arginine N-methyltransferase 8, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Lee, W.C, Ho, M.C. | | Deposit date: | 2014-12-02 | | Release date: | 2015-11-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Protein Arginine Methyltransferase 8: Tetrameric Structure and Protein Substrate Specificity
Biochemistry, 54, 2015
|
|
3TO6
 
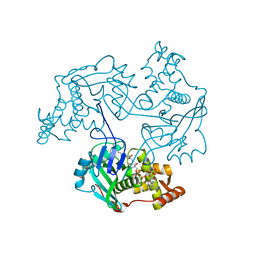 | |
3TO7
 
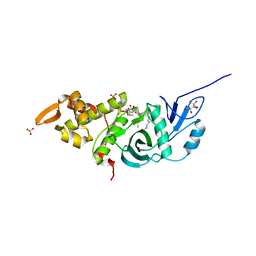 | | Crystal structure of yeast Esa1 HAT domain bound to coenzyme A with active site lysine acetylated | | Descriptor: | CACODYLIC ACID, COENZYME A, GLYCEROL, ... | | Authors: | Yuan, H, Ding, E.C, Marmorstein, R. | | Deposit date: | 2011-09-04 | | Release date: | 2011-11-09 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | MYST protein acetyltransferase activity requires active site lysine autoacetylation.
Embo J., 31, 2011
|
|
3TOB
 
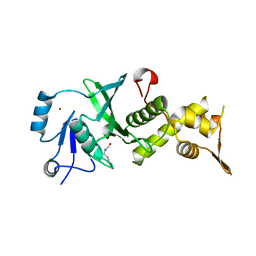 | |
3TO9
 
 | | Crystal structure of yeast Esa1 E338Q HAT domain bound to coenzyme A with active site lysine acetylated | | Descriptor: | 1,2-ETHANEDIOL, CACODYLIC ACID, COENZYME A, ... | | Authors: | Yuan, H, Ding, E.C, Marmorstein, R. | | Deposit date: | 2011-09-04 | | Release date: | 2011-11-09 | | Last modified: | 2012-01-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | MYST protein acetyltransferase activity requires active site lysine autoacetylation.
Embo J., 31, 2011
|
|
3TOA
 
 | | Human MOF crystal structure with active site lysine partially acetylated | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, ZINC ION, ... | | Authors: | Yuan, H, Ding, E.C, Marmorstein, R. | | Deposit date: | 2011-09-04 | | Release date: | 2011-11-09 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | MYST protein acetyltransferase activity requires active site lysine autoacetylation.
Embo J., 31, 2011
|
|
6BOZ
 
 | | Structure of human SETD8 in complex with covalent inhibitor MS4138 | | Descriptor: | 1,2-ETHANEDIOL, N-(3-{[7-(2-aminoethoxy)-6-methoxy-2-(pyrrolidin-1-yl)quinazolin-4-yl]amino}propyl)prop-2-enamide, N-lysine methyltransferase KMT5A | | Authors: | Babault, N, Anqi, M, Jin, J. | | Deposit date: | 2017-11-21 | | Release date: | 2019-05-01 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The dynamic conformational landscape of the protein methyltransferase SETD8.
Elife, 8, 2019
|
|
5UUT
 
 | | N-myristoyltransferase 1 (NMT) bound to myristoyl-CoA | | Descriptor: | CITRIC ACID, Glycylpeptide N-tetradecanoyltransferase 1, TETRADECANOYL-COA | | Authors: | Goodwin, O, Pegan, S. | | Deposit date: | 2017-02-17 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Blocking Myristoylation of Src Inhibits Its Kinase Activity and Suppresses Prostate Cancer Progression.
Cancer Res., 77, 2017
|
|
5V2N
 
 | | Crystal Structure of APO Human SETD8 | | Descriptor: | 1,2-ETHANEDIOL, N-lysine methyltransferase KMT5A | | Authors: | Skene, R.J. | | Deposit date: | 2017-03-05 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The dynamic conformational landscape of the protein methyltransferase SETD8.
Elife, 8, 2019
|
|
7WKQ
 
 | | Crystal Structure of halohydrin dehalogenase from Acidimicrobiia bacterium | | Descriptor: | halohydrin dehalogenase | | Authors: | Wan, N.W. | | Deposit date: | 2022-01-10 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Identification and Structure Analysis of an Unusual Halohydrin Dehalogenase for Highly Chemo-, Regio- and Enantioselective Bio-Nitration of Epoxides.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
