7Q5Y
 
 | |
1U6K
 
 | | TLS refinement of the structure of Se-methionine labelled Coenzyme f420-dependent methylenetetrahydromethanopterin dehydrogenase (MTD) from Methanopyrus kandleri | | Descriptor: | F420-dependent methylenetetrahydromethanopterin dehydrogenase, MAGNESIUM ION | | Authors: | Warkentin, E, Hagemeier, C.H, Shima, S, Thauer, R.K, Ermler, U. | | Deposit date: | 2004-07-30 | | Release date: | 2005-02-01 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The structure of F420-dependent methylenetetrahydromethanopterin dehydrogenase: a crystallographic 'superstructure' of the selenomethionine-labelled protein crystal structure.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1U6J
 
 | | The Structure of native coenzyme F420-dependent methylenetetrahydromethanopterin dehydrogenase at 2.4A resolution | | Descriptor: | F420-dependent methylenetetrahydromethanopterin dehydrogenase, MAGNESIUM ION | | Authors: | Warkentin, E, Hagemeier, C.H, Shima, S, Thauer, R.K, Ermler, U. | | Deposit date: | 2004-07-30 | | Release date: | 2005-02-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of F420-dependent methylenetetrahydromethanopterin dehydrogenase: a crystallographic 'superstructure' of the selenomethionine-labelled protein crystal structure.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1JAX
 
 | | Structure of Coenzyme F420H2:NADP+ Oxidoreductase (FNO) | | Descriptor: | MAGNESIUM ION, SODIUM ION, conserved hypothetical protein | | Authors: | Warkentin, E, Mamat, B, Thauer, R, Ermler, U, Shima, S. | | Deposit date: | 2001-06-01 | | Release date: | 2001-12-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of F420H2:NADP+ oxidoreductase with and without its substrates bound.
EMBO J., 20, 2001
|
|
1JAY
 
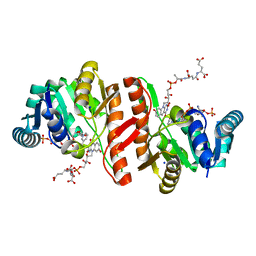 | | Structure of Coenzyme F420H2:NADP+ Oxidoreductase (FNO) with its substrates bound | | Descriptor: | COENZYME F420, Coenzyme F420H2:NADP+ Oxidoreductase (FNO), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Warkentin, E, Mamat, B, Thauer, R, Ermler, U, Shima, S. | | Deposit date: | 2001-06-01 | | Release date: | 2001-12-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of F420H2:NADP+ oxidoreductase with and without its substrates bound.
EMBO J., 20, 2001
|
|
1U6I
 
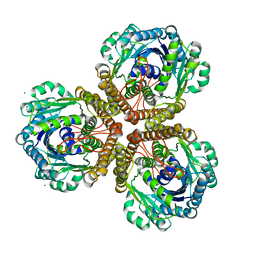 | | The Structure of native coenzyme F420-dependent methylenetetrahydromethanopterin dehydrogenase at 2.2A resolution | | Descriptor: | F420-dependent methylenetetrahydromethanopterin dehydrogenase, MAGNESIUM ION | | Authors: | Warkentin, E, Hagemeier, C.H, Shima, S, Thauer, R.K, Ermler, U. | | Deposit date: | 2004-07-30 | | Release date: | 2005-02-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of F420-dependent methylenetetrahydromethanopterin dehydrogenase: a crystallographic 'superstructure' of the selenomethionine-labelled protein crystal structure.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
5L9W
 
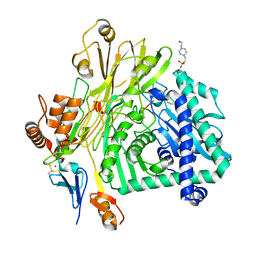 | | Crystal structure of the Apc core complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, Acetophenone carboxylase alpha subunit, ... | | Authors: | Warkentin, E, Weidenweber, S, Ermler, U. | | Deposit date: | 2016-06-11 | | Release date: | 2017-01-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the acetophenone carboxylase core complex: prototype of a new class of ATP-dependent carboxylases/hydrolases.
Sci Rep, 7, 2017
|
|
5OVQ
 
 | |
4S23
 
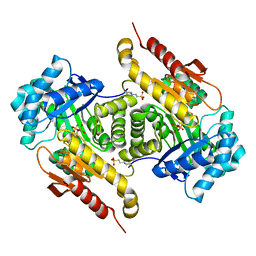 | | Structure of the GcpE-HMBPP complex from Thermus thermophilius | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, ... | | Authors: | Rekittke, I, Warkentin, E, Jomaa, H, Ermler, E. | | Deposit date: | 2015-01-19 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of the GcpE-HMBPP complex from Thermus thermophilius.
Biochem.Biophys.Res.Commun., 458, 2015
|
|
5DJQ
 
 | | The structure of CBB3 cytochrome oxidase. | | Descriptor: | CALCIUM ION, COPPER (II) ION, Cbb3-type cytochrome c oxidase subunit CcoN1, ... | | Authors: | Buschmann, S, Warkentin, E, Xie, H, Kohlstaedt, M, Langer, J.D, Ermler, U, Michel, H. | | Deposit date: | 2015-09-02 | | Release date: | 2016-01-13 | | Last modified: | 2016-07-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structure of cbb3 cytochrome oxidase provides insights into proton pumping.
Science, 329, 2010
|
|
2Y0F
 
 | | STRUCTURE OF GCPE (IspG) FROM THERMUS THERMOPHILUS HB27 | | Descriptor: | 4-HYDROXY-3-METHYLBUT-2-EN-1-YL DIPHOSPHATE SYNTHASE, IRON/SULFUR CLUSTER | | Authors: | Rekittke, I, Nonaka, T, Wiesner, J, Demmer, U, Warkentin, E, Jomaa, H, Ermler, U. | | Deposit date: | 2010-12-02 | | Release date: | 2011-01-26 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the E-1-Hydroxy-2-Methyl-But-2-Enyl-4-Diphosphate Synthase (Gcpe) from Thermus Thermophilus.
FEBS Lett., 585, 2011
|
|
2I2X
 
 | | Crystal structure of methanol:cobalamin methyltransferase complex MtaBC from Methanosarcina barkeri | | Descriptor: | 5-HYDROXYBENZIMIDAZOLYLCOB(III)AMIDE, Methyltransferase 1, POTASSIUM ION, ... | | Authors: | Hagemeier, C.H, Kruer, M, Thauer, R.K, Warkentin, E, Ermler, U. | | Deposit date: | 2006-08-17 | | Release date: | 2006-11-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insight into the mechanism of biological methanol activation based on the crystal structure of the methanol-cobalamin methyltransferase complex
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1EZW
 
 | | STRUCTURE OF COENZYME F420 DEPENDENT TETRAHYDROMETHANOPTERIN REDUCTASE FROM METHANOPYRUS KANDLERI | | Descriptor: | CHLORIDE ION, COENZYME F420-DEPENDENT N5,N10-METHYLENETETRAHYDROMETHANOPTERIN REDUCTASE, MAGNESIUM ION | | Authors: | Shima, S, Warkentin, E, Grabarse, W, Thauer, R.K, Ermler, U. | | Deposit date: | 2000-05-12 | | Release date: | 2000-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of coenzyme F(420) dependent methylenetetrahydromethanopterin reductase from two methanogenic archaea.
J.Mol.Biol., 300, 2000
|
|
1F07
 
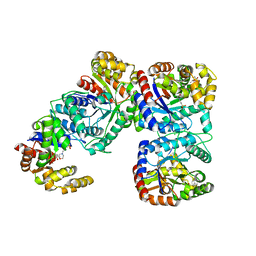 | | STRUCTURE OF COENZYME F420 DEPENDENT TETRAHYDROMETHANOPTERIN REDUCTASE FROM METHANOBACTERIUM THERMOAUTOTROPHICUM | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, ... | | Authors: | Shima, S, Warkentin, E, Grabarse, W, Thauer, R.K, Ermler, U. | | Deposit date: | 2000-05-15 | | Release date: | 2000-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of coenzyme F(420) dependent methylenetetrahydromethanopterin reductase from two methanogenic archaea.
J.Mol.Biol., 300, 2000
|
|
1Z47
 
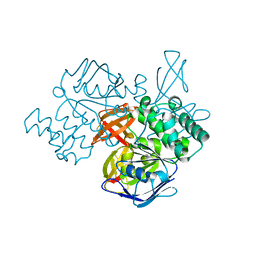 | | Structure of the ATPase subunit CysA of the putative sulfate ATP-binding cassette (ABC) transporter from Alicyclobacillus acidocaldarius | | Descriptor: | CHLORIDE ION, putative ABC-transporter ATP-binding protein | | Authors: | Scheffel, F, Demmer, U, Warkentin, E, Huelsmann, A, Schneider, E, Ermler, U. | | Deposit date: | 2005-03-15 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the ATPase subunit CysA of the putative sulfate ATP-binding cassette (ABC) transporter from Alicyclobacillus acidocaldarius
Febs Lett., 579, 2005
|
|
3F47
 
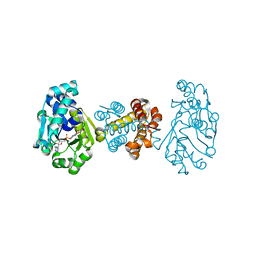 | | The Crystal Structure of [Fe]-Hydrogenase (Hmd) Holoenzyme from Methanocaldococcus jannaschii | | Descriptor: | 5'-O-[(S)-hydroxy{[2-hydroxy-3,5-dimethyl-6-(2-oxoethyl)pyridin-4-yl]oxy}phosphoryl]guanosine, 5,10-methenyltetrahydromethanopterin hydrogenase, CARBON MONOXIDE, ... | | Authors: | Hiromoto, T, Pilak, O, Warkentin, E, Thauer, R.K, Shima, S, Ermler, U. | | Deposit date: | 2008-10-31 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of C176A mutated [Fe]-hydrogenase suggests an acyl-iron ligation in the active site iron complex.
Febs Lett., 583, 2009
|
|
6TLK
 
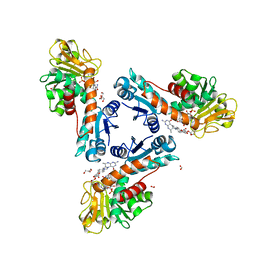 | | Structure of methylene-tetrahydromethanopterin dehydrogenase from Methylorubrum extorquens AM1 in an open conformation containing NADP+ and methylene-H4MPT | | Descriptor: | 5,10-DIMETHYLENE TETRAHYDROMETHANOPTERIN, Bifunctional protein MdtA, CHLORIDE ION, ... | | Authors: | Wagner, T, Huang, G, Demmer, U, Warkentin, E, Ermler, U, Shima, S. | | Deposit date: | 2019-12-02 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Hydride Transfer Process in NADP-dependent Methylene-tetrahydromethanopterin Dehydrogenase.
J.Mol.Biol., 432, 2020
|
|
6TM3
 
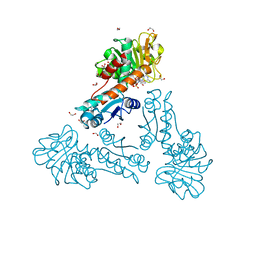 | | Structure of methylene-tetrahydromethanopterin dehydrogenase from Methylorubrum extorquens AM1 in a close conformation containing NADP+ and methylene-H4MPT | | Descriptor: | 1,2-ETHANEDIOL, 5,10-DIMETHYLENE TETRAHYDROMETHANOPTERIN, Bifunctional protein MdtA, ... | | Authors: | Wagner, T, Huang, G, Demmer, U, Warkentin, E, Ermler, U, Shima, S. | | Deposit date: | 2019-12-03 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | The Hydride Transfer Process in NADP-dependent Methylene-tetrahydromethanopterin Dehydrogenase.
J.Mol.Biol., 432, 2020
|
|
3OZV
 
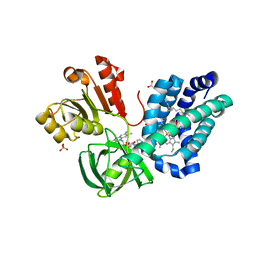 | | The Crystal Structure of flavohemoglobin from R. eutrophus in complex with econazole | | Descriptor: | 1-[(2S)-2-[(4-CHLOROBENZYL)OXY]-2-(2,4-DICHLOROPHENYL)ETHYL]-1H-IMIDAZOLE, 1-[GLYCEROLYLPHOSPHONYL]-2-[8-(2-HEXYL-CYCLOPROPYL)-OCTANAL-1-YL]-3-[HEXADECANAL-1-YL]-GLYCEROL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | El Hammi, E, Warkentin, E, Demmer, U, Ermler, U, Baciou, L. | | Deposit date: | 2010-09-27 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Ralstonia eutropha Flavohemoglobin in Complex with Three Antibiotic Azole Compounds.
Biochemistry, 50, 2011
|
|
3OZW
 
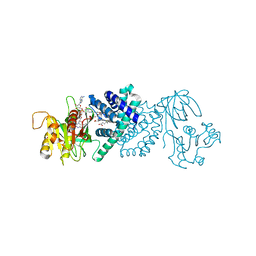 | | The Crystal Structure of flavohemoglobin from R. eutrophus in complex with ketoconazole | | Descriptor: | 1-[GLYCEROLYLPHOSPHONYL]-2-[8-(2-HEXYL-CYCLOPROPYL)-OCTANAL-1-YL]-3-[HEXADECANAL-1-YL]-GLYCEROL, 1-acetyl-4-(4-{[(2R,4S)-2-(2,4-dichlorophenyl)-2-(1H-imidazol-1-ylmethyl)-1,3-dioxolan-4-yl]methoxy}phenyl)piperazine, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | El Hammi, E, Warkentin, E, Demmer, U, Ermler, U, Baciou, L. | | Deposit date: | 2010-09-27 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Ralstonia eutropha Flavohemoglobin in Complex with Three Antibiotic Azole Compounds.
Biochemistry, 50, 2011
|
|
3OZU
 
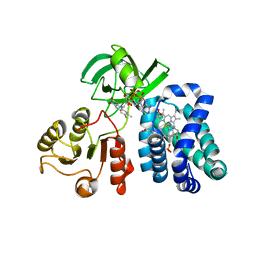 | | The Crystal Structure of flavohemoglobin from R. eutrophus in complex with miconazole | | Descriptor: | 1-[(2R)-2-[(2,4-dichlorobenzyl)oxy]-2-(2,4-dichlorophenyl)ethyl]-1H-imidazole, FLAVIN-ADENINE DINUCLEOTIDE, Flavohemoprotein, ... | | Authors: | El Hammi, E, Warkentin, E, Demmer, U, Ermler, U, Baciou, L. | | Deposit date: | 2010-09-27 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Ralstonia eutropha Flavohemoglobin in Complex with Three Antibiotic Azole Compounds.
Biochemistry, 50, 2011
|
|
4G1B
 
 | | X-ray structure of yeast flavohemoglobin in complex with econazole | | Descriptor: | 1-[(2S)-2-[(4-CHLOROBENZYL)OXY]-2-(2,4-DICHLOROPHENYL)ETHYL]-1H-IMIDAZOLE, FLAVIN-ADENINE DINUCLEOTIDE, Flavohemoglobin, ... | | Authors: | El Hammi, E, Warkentin, E, Demmer, U, Baciou, L, Ermler, U. | | Deposit date: | 2012-07-10 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Active site analysis of yeast flavohemoglobin based on its structure with a small ligand or econazole.
Febs J., 279, 2012
|
|
1SAU
 
 | | The Gamma subunit of the dissimilatory sulfite reductase (DsrC) from Archaeoglobus fulgidus at 1.1 A resolution | | Descriptor: | sulfite reductase, desulfoviridin-type subunit gamma | | Authors: | Mander, G.J, Weiss, M.S, Hedderich, R, Kahnt, J, Ermler, U, Warkentin, E. | | Deposit date: | 2004-02-09 | | Release date: | 2005-02-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Determination of a novel structure by a combination of long-wavelength sulfur phasing and radiation-damage-induced phasing.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1SB3
 
 | | Structure of 4-hydroxybenzoyl-CoA reductase from Thauera aromatica | | Descriptor: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-hydroxybenzoyl-CoA reductase alpha subunit, ... | | Authors: | Unciuleac, M, Warkentin, E, Page, C.C, Boll, M, Ermler, U. | | Deposit date: | 2004-02-10 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a Xanthine Oxidase-Related 4-Hydroxybenzoyl-CoA Reductase with an Additional [4Fe-4S] Cluster and an Inverted Electron Flow.
Structure, 12, 2004
|
|
1Z69
 
 | | Crystal structure of methylenetetrahydromethanopterin reductase (Mer) in complex with coenzyme F420 | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, CHLORIDE ION, COENZYME F420, ... | | Authors: | Aufhammer, S.W, Warkentin, E, Ermler, U, Hagemeier, C.H, Thauer, R.K, Shima, S. | | Deposit date: | 2005-03-22 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of methylenetetrahydromethanopterin reductase (Mer) in complex with coenzyme F420: Architecture of the F420/FMN binding site of enzymes within the nonprolyl cis-peptide containing bacterial luciferase family
Protein Sci., 14, 2005
|
|
