1ML1
 
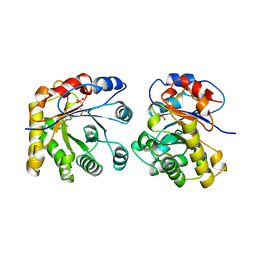 | | PROTEIN ENGINEERING WITH MONOMERIC TRIOSEPHOSPHATE ISOMERASE: THE MODELLING AND STRUCTURE VERIFICATION OF A SEVEN RESIDUE LOOP | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Thanki, N, Zeelen, J.P, Mathieu, M, Jaenicke, R, Abagyan, R.A, Wierenga, R, Schliebs, W. | | Deposit date: | 1996-09-27 | | Release date: | 1997-03-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Protein engineering with monomeric triosephosphate isomerase (monoTIM): the modelling and structure verification of a seven-residue loop.
Protein Eng., 10, 1997
|
|
1GAM
 
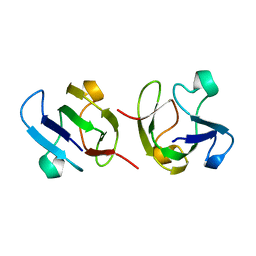 | | GAMMA B CRYSTALLIN TRUNCATED C-TERMINAL DOMAIN | | Descriptor: | GAMMA B CRYSTALLIN | | Authors: | Norledge, B.V, Mayr, E.-M, Glockshuber, R, Bateman, O.A, Slingsby, C, Jaenicke, R, Driessen, H.P.C. | | Deposit date: | 1996-02-02 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The X-ray structures of two mutant crystallin domains shed light on the evolution of multi-domain proteins.
Nat.Struct.Biol., 3, 1996
|
|
1NPS
 
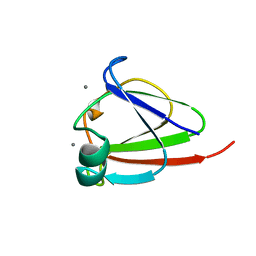 | | CRYSTAL STRUCTURE OF N-TERMINAL DOMAIN OF PROTEIN S | | Descriptor: | CALCIUM ION, DEVELOPMENT-SPECIFIC PROTEIN S | | Authors: | Wenk, M, Baumgartner, R, Mayer, E.M, Huber, R, Holak, T.A, Jaenicke, R. | | Deposit date: | 1999-02-01 | | Release date: | 2000-02-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The domains of protein S from Myxococcus xanthus: structure, stability and interactions.
J.Mol.Biol., 286, 1999
|
|
1VPE
 
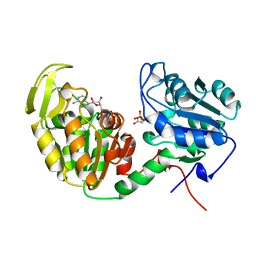 | | CRYSTALLOGRAPHIC ANALYSIS OF PHOSPHOGLYCERATE KINASE FROM THE HYPERTHERMOPHILIC BACTERIUM THERMOTOGA MARITIMA | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Auerbach, G, Huber, R, Graettinger, M, Zaiss, K, Schurig, H, Jaenicke, R, Jacob, U. | | Deposit date: | 1997-05-06 | | Release date: | 1998-06-17 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Closed structure of phosphoglycerate kinase from Thermotoga maritima reveals the catalytic mechanism and determinants of thermal stability.
Structure, 5, 1997
|
|
1HDG
 
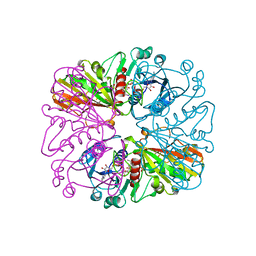 | | THE CRYSTAL STRUCTURE OF HOLO-GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE FROM THE HYPERTHERMOPHILIC BACTERIUM THERMOTOGA MARITIMA AT 2.5 ANGSTROMS RESOLUTION | | Descriptor: | HOLO-D-GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Korndoerfer, I, Steipe, B, Huber, R, Tomschy, A, Jaenicke, R. | | Deposit date: | 1995-01-17 | | Release date: | 1995-03-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of holo-glyceraldehyde-3-phosphate dehydrogenase from the hyperthermophilic bacterium Thermotoga maritima at 2.5 A resolution.
J.Mol.Biol., 246, 1995
|
|
1A5Z
 
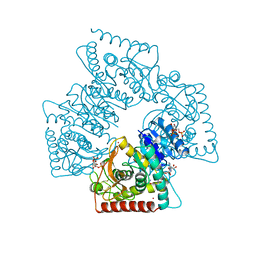 | | LACTATE DEHYDROGENASE FROM THERMOTOGA MARITIMA (TMLDH) | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, CADMIUM ION, L-LACTATE DEHYDROGENASE, ... | | Authors: | Auerbach, G, Ostendorp, R, Prade, L, Korndoerfer, I, Dams, T, Huber, R, Jaenicke, R. | | Deposit date: | 1998-02-18 | | Release date: | 1999-03-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Lactate dehydrogenase from the hyperthermophilic bacterium thermotoga maritima: the crystal structure at 2.1 A resolution reveals strategies for intrinsic protein stabilization.
Structure, 6, 1998
|
|
1DSL
 
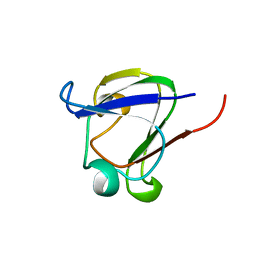 | | GAMMA B CRYSTALLIN C-TERMINAL DOMAIN | | Descriptor: | GAMMA B CRYSTALLIN | | Authors: | Norledge, B.V, Mayr, E.-M, Glockshuber, R, Bateman, O.A, Slingsby, C, Jaenicke, R, Driessen, H.P.C. | | Deposit date: | 1996-02-01 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The X-ray structures of two mutant crystallin domains shed light on the evolution of multi-domain proteins.
Nat.Struct.Biol., 3, 1996
|
|
1G6P
 
 | | SOLUTION NMR STRUCTURE OF THE COLD SHOCK PROTEIN FROM THE HYPERTHERMOPHILIC BACTERIUM THERMOTOGA MARITIMA | | Descriptor: | COLD SHOCK PROTEIN TMCSP | | Authors: | Kremer, W, Schuler, B, Harrieder, S, Geyer, M, Gronwald, W, Welker, C, Jaenicke, R, Kalbitzer, H.R. | | Deposit date: | 2000-11-07 | | Release date: | 2001-11-07 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the cold-shock protein from the hyperthermophilic bacterium Thermotoga maritima.
Eur.J.Biochem., 268, 2001
|
|
1HDF
 
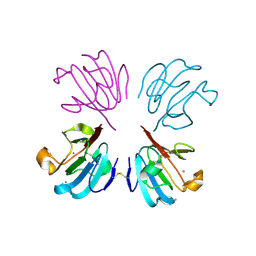 | | Evolution of the eye lens beta-gamma-crystallin domain fold | | Descriptor: | CALCIUM ION, SPHERULIN 3A | | Authors: | Clout, N.J, Kretschmar, M, Jaenicke, R, Slingsby, C. | | Deposit date: | 2000-11-13 | | Release date: | 2000-12-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of the Calcium-Loaded Spherulin 3A Dimer Sheds Light on the Evolution of the Eye Lens Betagamma-Crystallin Domain Fold
Structure, 9, 2001
|
|
1A5R
 
 | | STRUCTURE DETERMINATION OF THE SMALL UBIQUITIN-RELATED MODIFIER SUMO-1, NMR, 10 STRUCTURES | | Descriptor: | SUMO-1 | | Authors: | Bayer, P, Arndt, A, Metzger, S, Mahajan, R, Melchior, F, Jaenicke, R, Becker, J. | | Deposit date: | 1998-02-18 | | Release date: | 1998-10-14 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structure determination of the small ubiquitin-related modifier SUMO-1.
J.Mol.Biol., 280, 1998
|
|
1CZ3
 
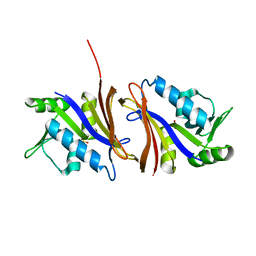 | | DIHYDROFOLATE REDUCTASE FROM THERMOTOGA MARITIMA | | Descriptor: | DIHYDROFOLATE REDUCTASE, SULFATE ION | | Authors: | Dams, T, Auerbach, G, Bader, G, Ploom, T, Huber, R, Jaenicke, R. | | Deposit date: | 1999-09-01 | | Release date: | 2000-03-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of dihydrofolate reductase from Thermotoga maritima: molecular features of thermostability.
J.Mol.Biol., 297, 2000
|
|
1D1G
 
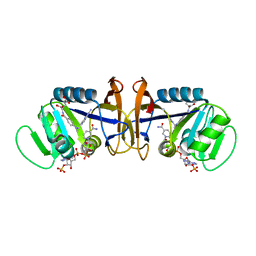 | | DIHYDROFOLATE REDUCTASE FROM THERMOTOGA MARITIMA | | Descriptor: | DIHYDROFOLATE REDUCTASE, METHOTREXATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Dams, T, Auerbach, G, Bader, G, Ploom, T, Huber, R, Jaenicke, R. | | Deposit date: | 1999-09-16 | | Release date: | 2000-03-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of dihydrofolate reductase from Thermotoga maritima: molecular features of thermostability.
J.Mol.Biol., 297, 2000
|
|
1A67
 
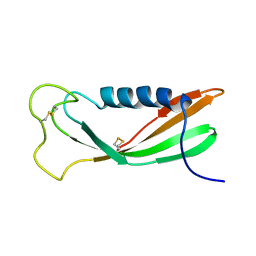 | | CHICKEN EGG WHITE CYSTATIN WILDTYPE, NMR, 16 STRUCTURES | | Descriptor: | CYSTATIN | | Authors: | Dieckmann, T, Mitschang, L, Hofmann, M, Kos, J, Turk, V, Auerswald, E.A, Jaenicke, R, Oschkinat, H. | | Deposit date: | 1998-03-06 | | Release date: | 1998-05-27 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The structures of native phosphorylated chicken cystatin and of a recombinant unphosphorylated variant in solution.
J.Mol.Biol., 234, 1993
|
|
1A90
 
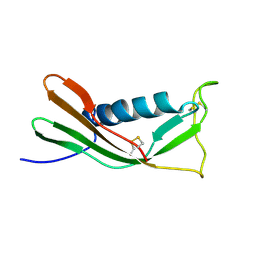 | | RECOMBINANT MUTANT CHICKEN EGG WHITE CYSTATIN, NMR, 31 STRUCTURES | | Descriptor: | CYSTATIN | | Authors: | Dieckmann, T, Mitschang, L, Hofmann, M, Kos, J, Turk, V, Auerswald, E.A, Jaenicke, R, Oschkinat, H. | | Deposit date: | 1998-04-14 | | Release date: | 1998-06-17 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The structures of native phosphorylated chicken cystatin and of a recombinant unphosphorylated variant in solution.
J.Mol.Biol., 234, 1993
|
|
1DIP
 
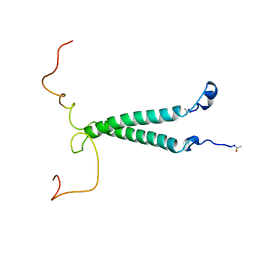 | | THE SOLUTION STRUCTURE OF PORCINE DELTA-SLEEP-INDUCING PEPTIDE IMMUNOREACTIVE PEPTIDE, NMR, 10 STRUCTURES | | Descriptor: | DELTA-SLEEP-INDUCING PEPTIDE IMMUNOREACTIVE PEPTIDE | | Authors: | Roesch, P, Seidel, G, Adermann, K, Schindler, T, Ejchart, A, Jaenicke, R, Forssmann, W.G. | | Deposit date: | 1997-04-09 | | Release date: | 1997-10-15 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of porcine delta sleep-inducing peptide immunoreactive peptide A homolog of the shortsighted gene product.
J.Biol.Chem., 272, 1997
|
|
1AG4
 
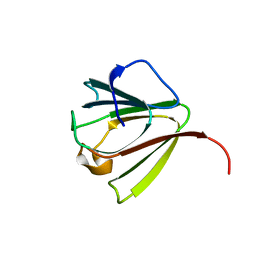 | | NMR STRUCTURE OF SPHERULIN 3A (S3A) FROM PHYSARUM POLYCEPHALUM, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | SPHERULIN 3A | | Authors: | Rosinke, B, Renner, C, Mayr, E.-M, Jaenicke, R, Holak, T.A. | | Deposit date: | 1997-04-01 | | Release date: | 1998-04-08 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Ca2+-loaded spherulin 3a from Physarum polycephalum adopts the prototype gamma-crystallin fold in aqueous solution.
J.Mol.Biol., 271, 1997
|
|
1E7N
 
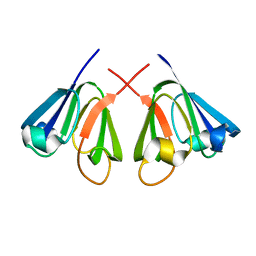 | | The N-terminal domain of beta-B2-crystallin resembles the putative ancestral homodimer | | Descriptor: | BETA-CRYSTALLIN B2 | | Authors: | Clout, N.J, Basak, A, Wieligmann, K, Bateman, O.A, Jaenicke, R, Slingsby, C. | | Deposit date: | 2000-08-31 | | Release date: | 2000-10-05 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The N-Terminal Domain of Betab2-Crystallin Resembles the Putative Ancestral Homodimer.
J.Mol.Biol., 304, 2000
|
|
1YTQ
 
 | |
1MSS
 
 | |
1TTJ
 
 | |
1TTI
 
 | |
1HPH
 
 | |
1CEW
 
 | |
1A7H
 
 | | GAMMA S CRYSTALLIN C-TERMINAL DOMAIN | | Descriptor: | GAMMAS CRYSTALLIN | | Authors: | Basak, A.K, Slingsby, C. | | Deposit date: | 1998-03-13 | | Release date: | 1998-05-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The C-terminal domains of gammaS-crystallin pair about a distorted twofold axis.
Protein Eng., 11, 1998
|
|
1B9B
 
 | | TRIOSEPHOSPHATE ISOMERASE OF THERMOTOGA MARITIMA | | Descriptor: | PROTEIN (TRIOSEPHOSPHATE ISOMERASE), SULFATE ION | | Authors: | Maes, D, Wierenga, R.K. | | Deposit date: | 1999-02-09 | | Release date: | 2000-01-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The crystal structure of triosephosphate isomerase (TIM) from Thermotoga maritima: a comparative thermostability structural analysis of ten different TIM structures.
Proteins, 37, 1999
|
|
