7F1W
 
 | | X-ray crystal structure of visual arrestin complexed with inositol hexaphosphate | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, S-arrestin | | Authors: | Kang, M, Jang, K, Eger, B.T, Ernst, O.P, Choe, H.W, Kim, Y.J. | | Deposit date: | 2021-06-10 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.097 Å) | | Cite: | Structural evidence for visual arrestin priming via complexation of phosphoinositols.
Structure, 30, 2022
|
|
7F1X
 
 | | X-ray crystal structure of visual arrestin complexed with inositol 1,4,5-triphosphate | | Descriptor: | 1,2-ETHANEDIOL, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, PENTANEDIAL, ... | | Authors: | Jang, K, Kang, M, Eger, B.T, Choe, H.W, Ernst, O.P, Kim, Y.J. | | Deposit date: | 2021-06-10 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural evidence for visual arrestin priming via complexation of phosphoinositols.
Structure, 30, 2022
|
|
4J4Q
 
 | | Crystal structure of active conformation of GPCR opsin stabilized by octylglucoside | | Descriptor: | ACETATE ION, Guanine nucleotide-binding protein G(t) subunit alpha-1, PALMITIC ACID, ... | | Authors: | Park, J.H, Morizumi, T, Li, Y, Hong, J.E, Pai, E.F, Hofmann, K.P, Choe, H.W, Ernst, O.P. | | Deposit date: | 2013-02-07 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Opsin, a structural model for olfactory receptors?
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
1RGC
 
 | | THE COMPLEX BETWEEN RIBONUCLEASE T1 AND 3'-GUANYLIC ACID SUGGESTS GEOMETRY OF ENZYMATIC REACTION PATH. AN X-RAY STUDY | | Descriptor: | CALCIUM ION, GUANOSINE-3'-MONOPHOSPHATE, RIBONUCLEASE T1 | | Authors: | Heydenreich, A, Koellner, G, Choe, H.W, Cordes, F, Kisker, C, Schindelin, H, Adamiak, R, Hahn, U, Saenger, W. | | Deposit date: | 1993-05-12 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The complex between ribonuclease T1 and 3'GMP suggests geometry of enzymic reaction path. An X-ray study.
Eur.J.Biochem., 218, 1993
|
|
3PQR
 
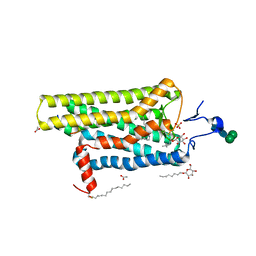 | | Crystal structure of Metarhodopsin II in complex with a C-terminal peptide derived from the Galpha subunit of transducin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Guanine nucleotide-binding protein G(t) subunit alpha-1, ... | | Authors: | Choe, H.-W, Kim, Y.J, Park, J.H, Morizumi, T, Pai, E.F, Krauss, N, Hofmann, K.P, Scheerer, P, Ernst, O.P. | | Deposit date: | 2010-11-26 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of metarhodopsin II.
Nature, 471, 2011
|
|
3PXO
 
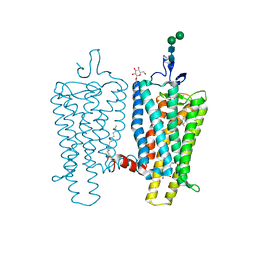 | | Crystal structure of Metarhodopsin II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITIC ACID, RETINAL, ... | | Authors: | Choe, H.-W, Kim, Y.J, Park, J.H, Morizumi, T, Pai, E.F, Krauss, N, Hofmann, K.P, Scheerer, P, Ernst, O.P. | | Deposit date: | 2010-12-10 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of metarhodopsin II.
Nature, 471, 2011
|
|
4BIR
 
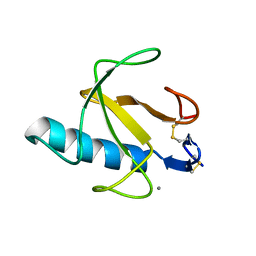 | | RIBONUCLEASE T1: FREE HIS92GLN MUTANT | | Descriptor: | CALCIUM ION, GUANYL-SPECIFIC RIBONUCLEASE T1 | | Authors: | Doumen, J, Steyaert, J. | | Deposit date: | 1998-01-13 | | Release date: | 1998-07-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Role of histidine-40 in ribonuclease T1 catalysis: three-dimensionalstructures of the partially active His40Lys mutant.
Biochemistry, 31, 1992
|
|
4RNT
 
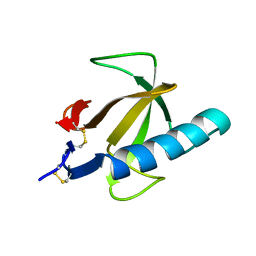 | |
9RNT
 
 | |
8RNT
 
 | | STRUCTURE OF RIBONUCLEASE T1 COMPLEXED WITH ZINC(II) AT 1.8 ANGSTROMS RESOLUTION: A ZN2+.6H2O.CARBOXYLATE CLATHRATE | | Descriptor: | RIBONUCLEASE T1, ZINC ION | | Authors: | Ding, J, Choe, H.-W, Granzin, J, Saenger, W. | | Deposit date: | 1991-09-23 | | Release date: | 1993-01-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of ribonuclease T1 complexed with zinc(II) at 1.8 A resolution: a Zn2+.6H2O.carboxylate clathrate.
Acta Crystallogr.,Sect.B, 48, 1992
|
|
1AYR
 
 | | ARRESTIN FROM BOVINE ROD OUTER SEGMENTS | | Descriptor: | ARRESTIN | | Authors: | Granzin, J, Wilden, U, Choe, H.-W, Labahn, J, Krafft, B, Bueldt, G. | | Deposit date: | 1997-11-10 | | Release date: | 1998-11-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | X-ray crystal structure of arrestin from bovine rod outer segments.
Nature, 391, 1998
|
|
2AAD
 
 | | THE ROLE OF HISTIDINE-40 IN RIBONUCLEASE T1 CATALYSIS: THREE-DIMENSIONAL STRUCTURES OF THE PARTIALLY ACTIVE HIS40LYS MUTANT | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE T1 ISOZYME | | Authors: | Zegers, I, Verhelst, P, Choe, C.W, Steyaert, J, Heinemann, U, Wyns, L, Saenger, W. | | Deposit date: | 1992-09-15 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of histidine-40 in ribonuclease T1 catalysis: three-dimensionalstructures of the partially active His40Lys mutant.
Biochemistry, 31, 1992
|
|
2AAE
 
 | | THE ROLE OF HISTIDINE-40 IN RIBONUCLEASE T1 CATALYSIS: THREE-DIMENSIONAL STRUCTURES OF THE PARTIALLY ACTIVE HIS40LYS MUTANT | | Descriptor: | CALCIUM ION, PHOSPHATE ION, RIBONUCLEASE T1 | | Authors: | Zegers, I, Verhelst, P, Choe, C.W, Steyaert, J, Heinemann, U, Wyns, L, Saenger, W. | | Deposit date: | 1992-09-15 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of histidine-40 in ribonuclease T1 catalysis: three-dimensionalstructures of the partially active His40Lys mutant.
Biochemistry, 31, 1992
|
|
4J2Q
 
 | |
3RNT
 
 | | CRYSTAL STRUCTURE OF GUANOSINE-FREE RIBONUCLEASE T1, COMPLEXED WITH VANADATE(V), SUGGESTS CONFORMATIONAL CHANGE UPON SUBSTRATE BINDING | | Descriptor: | CALCIUM ION, RIBONUCLEASE T1, VANADATE ION | | Authors: | Kostrewa, D, Choe, H.-W, Heinemann, U, Saenger, W. | | Deposit date: | 1989-05-31 | | Release date: | 1989-10-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of guanosine-free ribonuclease T1, complexed with vanadate (V), suggests conformational change upon substrate binding.
Biochemistry, 28, 1989
|
|
1LRA
 
 | | CRYSTALLOGRAPHIC STUDY OF GLU 58 ALA RNASE T1(ASTERISK)2'-GUANOSINE MONOPHOSPHATE AT 1.9 ANGSTROMS RESOLUTION | | Descriptor: | GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE T1, SODIUM ION | | Authors: | Pletinckx, J, Steyaert, J, Choe, H.-W, Heinemann, U, Wyns, L. | | Deposit date: | 1993-10-01 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic study of Glu58Ala RNase T1 x 2'-guanosine monophosphate at 1.9-A resolution.
Biochemistry, 33, 1994
|
|
5BIR
 
 | |
7JTB
 
 | |
7JSM
 
 | |
7JXA
 
 | | CRYSTAL STRUCTURE OF NATIVE BOVINE ARRESTIN 1 IN COMPLEX WITH INOSITOL 1,4,5-TRIPHOSPHATE | | Descriptor: | 2-ETHOXYETHANOL, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, S-arrestin, ... | | Authors: | Sander, C.L, Palczewski, K, Kiser, P.D. | | Deposit date: | 2020-08-26 | | Release date: | 2021-10-13 | | Last modified: | 2022-02-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural evidence for visual arrestin priming via complexation of phosphoinositols.
Structure, 30, 2022
|
|
1FIA
 
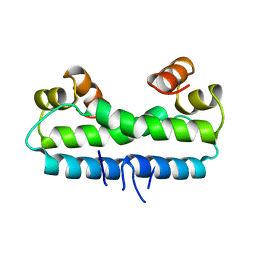 | | CRYSTAL STRUCTURE OF THE FACTOR FOR INVERSION STIMULATION FIS AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | FACTOR FOR INVERSION STIMULATION (FIS) | | Authors: | Kostrewa, D, Granzin, J, Choe, H.-W, Labahn, J, Saenger, W. | | Deposit date: | 1991-12-18 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the factor for inversion stimulation FIS at 2.0 A resolution.
J.Mol.Biol., 226, 1992
|
|
6R26
 
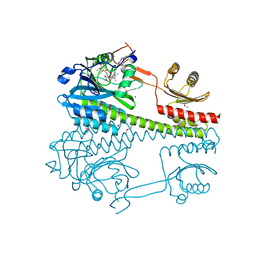 | | The photosensory core module (PAS-GAF-PHY) of the bacterial phytochrome Agp1 (AtBphP1) locked in a Pr-like state | | Descriptor: | 3-[2-[(~{Z})-[12-ethyl-6-(3-hydroxy-3-oxopropyl)-13-methyl-11-oxidanylidene-4,10-diazatricyclo[8.3.0.0^{3,7}]trideca-1,3,6,12-tetraen-5-ylidene]methyl]-5-[(~{Z})-(3-ethyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Bacteriophytochrome protein, CALCIUM ION | | Authors: | Scheerer, P, Michael, N, Lamparter, T, Krauss, N. | | Deposit date: | 2019-03-15 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Crystal structures of the photosensory core module of bacteriophytochrome Agp1 reveal pronounced structural flexibility of this protein in the red-absorbing Pr state
To Be Published
|
|
6R27
 
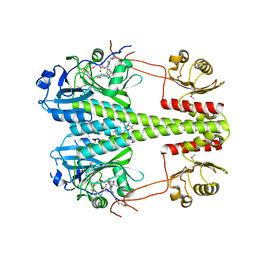 | | Crystallographic superstructure of the photosensory core module (PAS-GAF-PHY) of the bacterial phytochrome Agp1 (AtBphP1) locked in a Pr-like state | | Descriptor: | 3-[2-[(~{Z})-[12-ethyl-6-(3-hydroxy-3-oxopropyl)-13-methyl-11-oxidanylidene-4,10-diazatricyclo[8.3.0.0^{3,7}]trideca-1,3,6,12-tetraen-5-ylidene]methyl]-5-[(~{Z})-(3-ethyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Bacteriophytochrome protein | | Authors: | Scheerer, P, Michael, N, Lamparter, T, Krauss, N. | | Deposit date: | 2019-03-15 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structures of the photosensory core module of bacteriophytochrome Agp1 reveal pronounced structural flexibility of this protein in the red-absorbing Pr state
To Be Published
|
|
7MP2
 
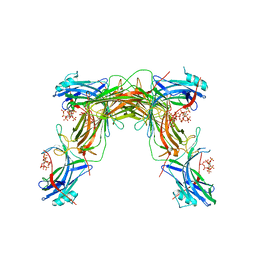 | | CRYSTAL STRUCTURE OF NATIVE BOVINE ARRESTIN 1 IN COMPLEX WITH 1D-MYO-INOSITOL 1,5-BISDIPHOSPHATE TETRAKISPHOSPHATE (1,5-PP IP4) | | Descriptor: | (1R,3S,4R,5S,6R)-2,4,5,6-tetrakis(phosphonooxy)cyclohexane-1,3-diyl bis[trihydrogen (diphosphate)], S-arrestin | | Authors: | Sander, C.L, Palczewski, K, Kiser, P.D. | | Deposit date: | 2021-05-04 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural evidence for visual arrestin priming via complexation of phosphoinositols.
Structure, 30, 2022
|
|
7MP0
 
 | | CRYSTAL STRUCTURE OF NATIVE BOVINE ARRESTIN 1 IN COMPLEX WITH 1D-MYO-INOSITOL 5-DIPHOSPHATE PENTAKISPHOSPHATE (5-PP IP5) | | Descriptor: | (1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, 2-ETHOXYETHANOL, S-arrestin | | Authors: | Sander, C.L, Palczewski, K, Kiser, P.D. | | Deposit date: | 2021-05-04 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural evidence for visual arrestin priming via complexation of phosphoinositols.
Structure, 30, 2022
|
|
