1APA
 
 | | X-RAY STRUCTURE OF A POKEWEED ANTIVIRAL PROTEIN, CODED BY A NEW GENOMIC CLONE, AT 0.23 NM RESOLUTION. A MODEL STRUCTURE PROVIDES A SUITABLE ELECTROSTATIC FIELD FOR SUBSTRATE BINDING. | | Descriptor: | POKEWEED ANTIVIRAL PROTEIN | | Authors: | Ago, H, Kataoka, J, Tsuge, H, Habuka, N, Inagaki, E, Noma, M, Miyano, M. | | Deposit date: | 1993-09-21 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray structure of a pokeweed antiviral protein, coded by a new genomic clone, at 0.23 nm resolution. A model structure provides a suitable electrostatic field for substrate binding.
Eur.J.Biochem., 225, 1994
|
|
1CIW
 
 | | PEANUT LECTIN COMPLEXED WITH N-ACETYLLACTOSAMINE | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, PROTEIN (PEANUT LECTIN), ... | | Authors: | Ravishankar, R, Suguna, K, Surolia, A, Vijayan, M. | | Deposit date: | 1999-04-06 | | Release date: | 1999-07-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of the complexes of peanut lectin with methyl-beta-galactose and N-acetyllactosamine and a comparative study of carbohydrate binding in Gal/GalNAc-specific legume lectins.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1LQG
 
 | | ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE COMPLEX WITH URACIL-DNA GLYCOSYLASE INHIBITOR PROTEIN | | Descriptor: | URACIL-DNA GLYCOSYLASE, URACIL-DNA GLYCOSYLASE INHIBITOR | | Authors: | Saikrishnan, K, Sagar, M.B, Ravishankar, R, Roy, S, Purnapatre, K, Handa, P, Varshney, U, Vijayan, M. | | Deposit date: | 2002-05-10 | | Release date: | 2002-11-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Domain closure and action of uracil DNA glycosylase (UDG): structures of new crystal forms containing the Escherichia coli enzyme and a comparative study of the known structures involving UDG.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1QAW
 
 | | Regulatory Features of the TRP Operon and the Crystal Structure of the TRP RNA-Binding Attenuation Protein from Bacillus Stearothermophilus. | | Descriptor: | TRP RNA-BINDING ATTENUATION PROTEIN, TRYPTOPHAN | | Authors: | Chen, X.-P, Antson, A.A, Yang, M, Baumann, C, Dodson, E.J, Dodson, G.G, Gollnick, P. | | Deposit date: | 1999-03-31 | | Release date: | 1999-04-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Regulatory features of the trp operon and the crystal structure of the trp RNA-binding attenuation protein from Bacillus stearothermophilus.
J.Mol.Biol., 289, 1999
|
|
1LQJ
 
 | | ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE | | Descriptor: | URACIL-DNA GLYCOSYLASE | | Authors: | Saikrishnan, K, Sagar, M.B, Ravishankar, R, Roy, S, Purnapatre, K, Varshney, U, Vijayan, M. | | Deposit date: | 2002-05-10 | | Release date: | 2002-11-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Domain closure and action of uracil DNA glycosylase (UDG): structures of new crystal forms containing the Escherichia coli enzyme and a comparative study of the known structures involving UDG.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1LQM
 
 | | ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE COMPLEX WITH URACIL-DNA GLYCOSYLASE INHIBITOR PROTEIN | | Descriptor: | URACIL-DNA GLYCOSYLASE, URACIL-DNA GLYCOSYLASE INHIBITOR | | Authors: | Saikrishnan, K, Sagar, M.B, Ravishankar, R, Roy, S, Purnapatre, K, Varshney, U, Vijayan, M. | | Deposit date: | 2002-05-10 | | Release date: | 2002-11-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Domain closure and action of uracil DNA glycosylase (UDG): structures of new crystal forms containing the Escherichia coli enzyme and a comparative study of the known structures involving UDG.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1Y8I
 
 | | Horse methemoglobin low salt, PH 7.0 (98% relative humidity) | | Descriptor: | Hemoglobin alpha chains, Hemoglobin beta chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sankaranarayanan, R, Biswal, B.K, Vijayan, M. | | Deposit date: | 2004-12-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A new relaxed state in horse methemoglobin characterized by crystallographic studies.
Proteins, 60, 2005
|
|
1Y8K
 
 | | Horse methemoglobin low salt, PH 7.0 (88% relative humidity) | | Descriptor: | Hemoglobin alpha chains, Hemoglobin beta chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sankaranarayanan, R, Biswal, B.K, Vijayan, M. | | Deposit date: | 2004-12-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A new relaxed state in horse methemoglobin characterized by crystallographic studies.
Proteins, 60, 2005
|
|
1XEI
 
 | |
1XEK
 
 | |
1XEJ
 
 | |
7VX3
 
 | | OXA-58 crystal structure of acylated meropenem complex 2 | | Descriptor: | (2S,3R,4S)-4-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase, ... | | Authors: | Saino, H, Sugiyabu, T, Miyano, M. | | Deposit date: | 2021-11-12 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | OXA-58 crystal structure of acylated meropenem complex 2
To be published
|
|
7VX6
 
 | | OXA-58 crystal structure of acylated meropenem complex 2 | | Descriptor: | (2S,3R,4S)-4-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase, ... | | Authors: | Saino, H, Sugiyabu, T, Miyano, M. | | Deposit date: | 2021-11-12 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | OXA-58 crystal structure of acylated meropenem complex 2
To be published
|
|
1Y8H
 
 | | HORSE METHEMOGLOBIN LOW SALT, PH 7.0 | | Descriptor: | Hemoglobin alpha chains, Hemoglobin beta chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sankaranarayanan, R, Biswal, B.K, Vijayan, M. | | Deposit date: | 2004-12-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A new relaxed state in horse methemoglobin characterized by crystallographic studies.
Proteins, 60, 2005
|
|
3IQU
 
 | |
1LJ4
 
 | |
1LJI
 
 | |
3IQJ
 
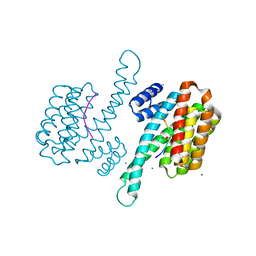 | | Crystal Structure of human 14-3-3 sigma in Complex with Raf1 peptide (10mer) | | Descriptor: | 10-mer peptide from RAF proto-oncogene serine/threonine-protein kinase, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Ottmann, C, Weyand, M. | | Deposit date: | 2009-08-20 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Impaired binding of 14-3-3 to C-RAF in Noonan syndrome suggests new approaches in diseases with increased Ras signaling.
Mol.Cell.Biol., 30, 2010
|
|
1LJJ
 
 | |
1LJE
 
 | |
1LJF
 
 | |
1LJ3
 
 | |
1LJK
 
 | |
1JAC
 
 | | A NOVEL MODE OF CARBOHYDRATE RECOGNITION IN JACALIN, A MORACEAE PLANT LECTIN WITH A BETA-PRISM | | Descriptor: | JACALIN, methyl alpha-D-galactopyranoside | | Authors: | Sankaranarayanan, R, Sekar, K, Banerjee, R, Sharma, V, Surolia, A, Vijayan, M. | | Deposit date: | 1996-05-22 | | Release date: | 1997-06-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | A novel mode of carbohydrate recognition in jacalin, a Moraceae plant lectin with a beta-prism fold.
Nat.Struct.Biol., 3, 1996
|
|
1LJH
 
 | |
