6AP7
 
 | |
6AP8
 
 | |
6AP6
 
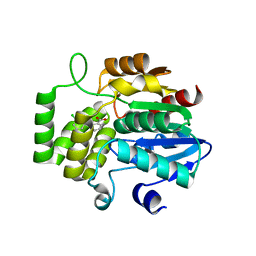 | | Crystal Structure of DAD2 in complex with tolfenamic acid | | Descriptor: | 2-[(3-chloro-2-methylphenyl)amino]benzoic acid, Probable strigolactone esterase DAD2 | | Authors: | Hamiaux, C. | | Deposit date: | 2017-08-17 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Inhibition of strigolactone receptors byN-phenylanthranilic acid derivatives: Structural and functional insights.
J. Biol. Chem., 293, 2018
|
|
8ITR
 
 | |
8ITT
 
 | |
7F6L
 
 | | Crystal structure of human MUS81-EME2 complex | | Descriptor: | Crossover junction endonuclease MUS81, Probable crossover junction endonuclease EME2 | | Authors: | Hua, Z.K, Zhang, D.P, Yuan, C, Lin, Z.H. | | Deposit date: | 2021-06-25 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the human MUS81-EME2 complex.
Structure, 30, 2022
|
|
8J66
 
 | | Crystal structure of glycosyltransferase SgUGT94-289-3 in complex with M3, state 2 | | Descriptor: | (2S,3S,4S,5R,6R)-2-(hydroxymethyl)-6-[[(2R,3S,4S,5R,6S)-6-[(3R,6S)-6-[(3S,8S,9R,10R,11S,13R,14S,17S)-3-[(2R,3R,4S,5S,6S)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-4,4,9,13,14-pentamethyl-11-oxidanyl-2,3,7,8,10,11,12,15,16,17-decahydro-1H-cyclopenta[a]phenanthren-17-yl]-2-methyl-2-oxidanyl-heptan-3-yl]oxy-3,4,5-tris(oxidanyl)oxan-2-yl]methoxy]oxane-3,4,5-triol, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Li, M, Zhang, S, Cui, S. | | Deposit date: | 2023-04-24 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural insights into the catalytic selectivity of glycosyltransferase SgUGT94-289-3 towards mogrosides.
Nat Commun, 15, 2024
|
|
7F5H
 
 | | The crystal structure of RBD-Nanobody complex, DL28 (SC4) | | Descriptor: | GLYCEROL, Nanobody DL28, PHOSPHATE ION, ... | | Authors: | Luo, Z.P, Li, T, Lai, Y, Zhou, Y, Tan, J, Li, D. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Characterization of a Neutralizing Nanobody With Broad Activity Against SARS-CoV-2 Variants.
Front Microbiol, 13, 2022
|
|
7WMI
 
 | | Threonyl-tRNA synthetase from Salmonella enterica in complex with an inhibitor | | Descriptor: | (2S,3R)-2-azanyl-N-[(1R)-2-[3-(7-bromanyl-6-chloranyl-4-oxidanylidene-quinazolin-3-yl)propanoylamino]-1-(3-phenylphenyl)ethyl]-3-oxidanyl-butanamide, 1,2-ETHANEDIOL, Threonine--tRNA ligase, ... | | Authors: | Cai, Z, Chen, B, Yu, Y, Zhou, H. | | Deposit date: | 2022-01-14 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, Synthesis, and Proof-of-Concept of Triple-Site Inhibitors against Aminoacyl-tRNA Synthetases.
J.Med.Chem., 65, 2022
|
|
7FIA
 
 | | Structure of AcrIF23 | | Descriptor: | AcrIF23 | | Authors: | Ren, J, Yue, F. | | Deposit date: | 2021-07-30 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural and mechanistic insights into the inhibition of type I-F CRISPR-Cas system by anti-CRISPR protein AcrIF23.
J.Biol.Chem., 298, 2022
|
|
7VM8
 
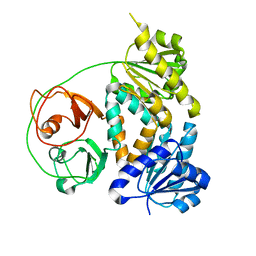 | | Crystal structure of the MtDMI1 gating ring | | Descriptor: | Ion channel DMI1 | | Authors: | Huang, X, Zhang, P. | | Deposit date: | 2021-10-08 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.034 Å) | | Cite: | Constitutive activation of a nuclear-localized calcium channel complex in Medicago truncatula.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7YC2
 
 | |
7EIV
 
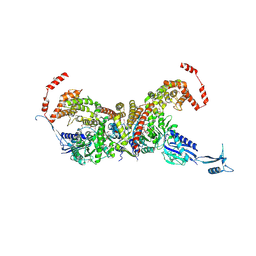 | | heterotetrameric glycyl-tRNA synthetase from Escherichia coli | | Descriptor: | GLYCINE, Glycine--tRNA ligase alpha subunit, Glycine--tRNA ligase beta subunit, ... | | Authors: | Ju, Y, Zhou, H. | | Deposit date: | 2021-03-31 | | Release date: | 2021-08-11 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | X-shaped structure of bacterial heterotetrameric tRNA synthetase suggests cryptic prokaryote functions and a rationale for synthetase classifications.
Nucleic Acids Res., 49, 2021
|
|
7YSE
 
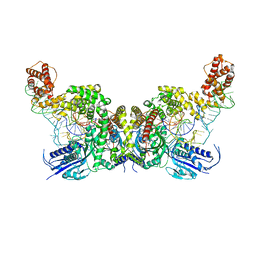 | | Crystal structure of E. coli heterotetrameric GlyRS in complex with tRNA | | Descriptor: | Glycine--tRNA ligase alpha subunit, Glycine--tRNA ligase beta subunit, MAGNESIUM ION, ... | | Authors: | Han, L, Ju, Y, Zhou, H. | | Deposit date: | 2022-08-12 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.907 Å) | | Cite: | The binding mode of orphan glycyl-tRNA synthetase with tRNA supports the synthetase classification and reveals large domain movements.
Sci Adv, 9, 2023
|
|
7F5G
 
 | | The crystal structure of RBD-Nanobody complex, DL4 (SA4) | | Descriptor: | ACETATE ION, GLYCEROL, Nanobody DL4, ... | | Authors: | Li, T, Lai, Y, Zhou, Y, Tan, J, Li, D. | | Deposit date: | 2021-06-22 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Isolation, characterization, and structure-based engineering of a neutralizing nanobody against SARS-CoV-2.
Int.J.Biol.Macromol., 209, 2022
|
|
4I8B
 
 | | Crystal Structure of Thioredoxin from Schistosoma Japonicum | | Descriptor: | Thioredoxin | | Authors: | Wu, Q, Peng, Y, Zhao, J, Li, X, Fan, X, Zhou, X, Chen, J, Luo, Z, Shi, D. | | Deposit date: | 2012-12-03 | | Release date: | 2013-12-04 | | Last modified: | 2015-06-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Expression, characterization and crystal structure of thioredoxin from Schistosoma japonicum.
Parasitology, 142, 2015
|
|
6LI1
 
 | | Crystal structure of GPR52 ligand free form with flavodoxin fusion | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Chimera of G-protein coupled receptor 52 and Flavodoxin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Luo, Z.P, Lin, X, Xu, F, Han, G.W. | | Deposit date: | 2019-12-10 | | Release date: | 2020-02-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of ligand recognition and self-activation of orphan GPR52.
Nature, 579, 2020
|
|
6LI0
 
 | | Crystal structure of GPR52 in complex with agonist c17 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CITRATE ANION, Chimera of G-protein coupled receptor 52 and Flavodoxin, ... | | Authors: | Luo, Z.P, Lin, X, Xu, F, Han, G.W. | | Deposit date: | 2019-12-10 | | Release date: | 2020-02-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of ligand recognition and self-activation of orphan GPR52.
Nature, 579, 2020
|
|
6LI2
 
 | | Crystal structure of GPR52 ligand free form with rubredoxin fusion | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Chimera of G-protein coupled receptor 52 and Rubredoxin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Luo, Z.P, Lin, X, Xu, F, Han, G.W. | | Deposit date: | 2019-12-10 | | Release date: | 2020-02-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of ligand recognition and self-activation of orphan GPR52.
Nature, 579, 2020
|
|
6LI3
 
 | | cryo-EM structure of GPR52-miniGs-NB35 | | Descriptor: | G-protein coupled receptor 52, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Li, M, Wang, N, Xu, F, Wu, J, Lei, M. | | Deposit date: | 2019-12-10 | | Release date: | 2020-02-26 | | Last modified: | 2020-03-18 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structural basis of ligand recognition and self-activation of orphan GPR52.
Nature, 579, 2020
|
|
