3KFO
 
 | | Crystal structure of the C-terminal domain from the nuclear pore complex component NUP133 from Saccharomyces cerevisiae | | Descriptor: | GLYCEROL, Nucleoporin NUP133 | | Authors: | Sampathkumar, P, Bonanno, J.B, Miller, S, Bain, K, Dickey, M, Gheyi, T, Almo, S.C, Rout, M, Sali, A, Phillips, J, Pieper, U, Fernandez-Martinez, J, Franke, J.D, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-27 | | Release date: | 2010-01-26 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the C-terminal domain of Saccharomyces cerevisiae Nup133, a component of the nuclear pore complex.
Proteins, 79, 2011
|
|
2MAS
 
 | | PURINE NUCLEOSIDE HYDROLASE WITH A TRANSITION STATE INHIBITOR | | Descriptor: | 2-(4-AMINO-PHENYL)-5-HYDROXYMETHYL-PYRROLIDINE-3,4-DIOL, CALCIUM ION, INOSINE-URIDINE NUCLEOSIDE N-RIBOHYDROLASE | | Authors: | Degano, M, Schramm, V.L, Sacchettini, J.C. | | Deposit date: | 1996-10-17 | | Release date: | 1997-08-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Trypanosomal nucleoside hydrolase. A novel mechanism from the structure with a transition-state inhibitor.
Biochemistry, 37, 1998
|
|
2N8L
 
 | |
2N8M
 
 | |
2PGE
 
 | |
2QUX
 
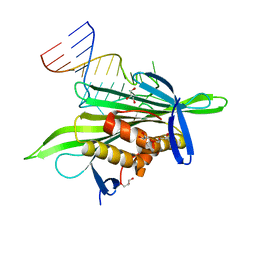 | | PP7 coat protein dimer in complex with RNA hairpin | | Descriptor: | Coat protein, GLYCEROL, RNA (25-MER) | | Authors: | Chao, J.A. | | Deposit date: | 2007-08-06 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural basis for the coevolution of a viral RNA-protein complex.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2QUD
 
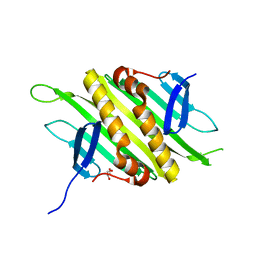 | | PP7 Coat Protein Dimer | | Descriptor: | Coat protein, GLYCEROL | | Authors: | Chao, J.A. | | Deposit date: | 2007-08-04 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the coevolution of a viral RNA-protein complex.
Nat.Struct.Mol.Biol., 15, 2008
|
|
1YVN
 
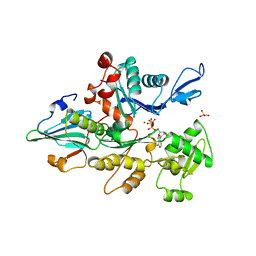 | | THE YEAST ACTIN VAL 159 ASN MUTANT COMPLEX WITH HUMAN GELSOLIN SEGMENT 1. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Vorobiev, S.M, Belmont, L.D, Drubin, D.G, Almo, S.C. | | Deposit date: | 1999-03-23 | | Release date: | 2000-03-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of yeast actin V159N mutant
To be Published
|
|
3RIT
 
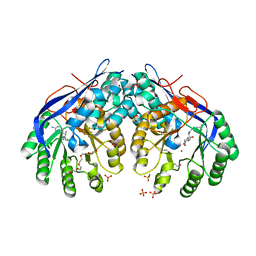 | | Crystal structure of Dipeptide Epimerase from Methylococcus capsulatus complexed with Mg and dipeptide L-Arg-D-Lys | | Descriptor: | ARGININE, D-LYSINE, Dipeptide epimerase, ... | | Authors: | Lukk, T, Sakai, A, Song, L, Gerlt, J.A, Nair, S.K. | | Deposit date: | 2011-04-14 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3RO6
 
 | | Crystal structure of Dipeptide Epimerase from Methylococcus capsulatus complexed with Mg ion | | Descriptor: | GLYCEROL, MAGNESIUM ION, Putative chloromuconate cycloisomerase, ... | | Authors: | Lukk, T, Sakai, A, Song, L, Gerlt, J.A, Nair, S.K. | | Deposit date: | 2011-04-25 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3FOW
 
 | | Plasmodium Purine Nucleoside Phosphorylase V66I-V73I-Y160F Mutant | | Descriptor: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, PHOSPHATE ION, Uridine phosphorylase, ... | | Authors: | Donaldson, T, Zhan, C. | | Deposit date: | 2009-01-02 | | Release date: | 2009-12-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural determinants of the 5'-methylthioinosine specificity of Plasmodium purine nucleoside phosphorylase.
Plos One, 9, 2014
|
|
3F1C
 
 | | CRYSTAL STRUCTURE OF 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase from Listeria monocytogenes | | Descriptor: | Putative 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase 2 | | Authors: | Patskovsky, Y, Ho, J, Toro, R, Gilmore, M, Miller, S, Groshong, C, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-27 | | Release date: | 2008-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CRYSTAL STRUCTURE OF 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase from Listeria monocytogenes
To be Published
|
|
1ASF
 
 | |
1ASG
 
 | |
3B9X
 
 | |
3AAT
 
 | |
1ASC
 
 | |
1ASB
 
 | |
1ASD
 
 | |
1ASE
 
 | |
1YOE
 
 | |
1XFT
 
 | |
2AI2
 
 | | Purine nucleoside phosphorylase from calf spleen | | Descriptor: | ((2S,3AS,4R,6S)-4-(HYDROXYMETHYL)-6-(4-OXO-4,5-DIHYDRO-3H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)-TETRAHYDROFURO[3,4-D][1,3]DIOXO L-2-YL)METHYLPHOSPHONIC ACID, MAGNESIUM ION, Purine nucleoside phosphorylase, ... | | Authors: | Toms, A.V, Wang, W, Li, Y, Ganem, B, Ealick, S.E. | | Deposit date: | 2005-07-28 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Novel multisubstrate inhibitors of mammalian purine nucleoside phosphorylase.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2AI3
 
 | | Purine nucleoside phosphorylase from calf spleen | | Descriptor: | (2S,4R,6R,6AS)-4-(2-AMINO-6-OXO-1,6-DIHYDROPURIN-9-YL)-6-(HYDROXYMETHYL)-TETRAHYDROFURO[3,4-D][1,3]DIOXOL-2-YLPHOSPHONI C ACID, MAGNESIUM ION, Purine nucleoside phosphorylase, ... | | Authors: | Toms, A.V, Wang, W, Li, Y, Ganem, B, Ealick, S.E. | | Deposit date: | 2005-07-28 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Novel multisubstrate inhibitors of mammalian purine nucleoside phosphorylase.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2AJT
 
 | | Crystal structure of L-Arabinose Isomerase from E.coli | | Descriptor: | L-arabinose isomerase | | Authors: | Manjasetty, B.A, Fedorov, E.V, Almo, S.C, Chance, M.R, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-08-02 | | Release date: | 2005-08-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Escherichia coli L-Arabinose Isomerase (ECAI), The Putative Target of Biological Tagatose Production
J.Mol.Biol., 360, 2006
|
|
