8GA7
 
 | |
8G9J
 
 | |
7PB9
 
 | | Crystal structure of tandem WH domains of Vps25 from Odinarchaeota | | Descriptor: | Tandem WH domains of Vps25 | | Authors: | Salzer, R, Bellini, D, Papatziamou, D, Robinson, N.P, Lowe, J. | | Deposit date: | 2021-08-01 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Asgard archaea shed light on the evolutionary origins of the eukaryotic ubiquitin-ESCRT machinery.
Nat Commun, 13, 2022
|
|
5KAF
 
 | | RT XFEL structure of Photosystem II in the dark state at 3.0 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Fuller, F, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Waterman, D.G, Evans, G, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-06-01 | | Release date: | 2016-11-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.00001 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
5KAI
 
 | | NH3-bound RT XFEL structure of Photosystem II 500 ms after the 2nd illumination (2F) at 2.8 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Fuller, F, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Waterman, D.G, Evans, G, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-06-01 | | Release date: | 2016-11-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.80000925 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
6O0C
 
 | | NMR ensemble of computationally designed protein XAA_GVDQ mutant M4L | | Descriptor: | Design construct XAA_GVDQ mutant M4L | | Authors: | Wei, K.Y, Moschidi, D, Nerli, S, Sgourakis, N, Baker, D. | | Deposit date: | 2019-02-15 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Computational design of closely related proteins that adopt two well-defined but structurally divergent folds.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6BFT
 
 | | Structure of Bevacizumab Fab mutant in complex with VEGF | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Avastin Heavy Chain Fab fragment mutant, Avastin Light Chain Fab fragment mutant, ... | | Authors: | Christie, M, Christ, D. | | Deposit date: | 2017-10-27 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Stable human IgG antibody therapeutics with native
framework structure
To Be Published
|
|
6N3R
 
 | |
8FYT
 
 | | LSD-bound serotonin 1A (5-HT1A) receptor-Gi1 protein complex | | Descriptor: | (8alpha)-N,N-diethyl-6-methyl-9,10-didehydroergoline-8-carboxamide, CHOLESTEROL HEMISUCCINATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Warren, A.L, Zilberg, G, Capper, M.J, Wacker, D. | | Deposit date: | 2023-01-26 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Structural pharmacology and therapeutic potential of 5-methoxytryptamines.
Nature, 630, 2024
|
|
8FYL
 
 | | Vilazodone-bound serotonin 1A (5-HT1A) receptor-Gi1 protein complex | | Descriptor: | 5-{4-[4-(5-cyano-1H-indol-3-yl)butyl]piperazin-1-yl}-1-benzofuran-2-carboxamide, CHOLESTEROL HEMISUCCINATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Warren, A.L, Zilberg, G, Capper, M.J, Wacker, D. | | Deposit date: | 2023-01-26 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural pharmacology and therapeutic potential of 5-methoxytryptamines.
Nature, 630, 2024
|
|
8FYX
 
 | | Buspirone-bound serotonin 1A (5-HT1A) receptor-Gi1 protein complex | | Descriptor: | Buspirone, CHOLESTEROL HEMISUCCINATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Warren, A.L, Zilberg, G, Capper, M.J, Wacker, D. | | Deposit date: | 2023-01-26 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Structural pharmacology and therapeutic potential of 5-methoxytryptamines.
Nature, 630, 2024
|
|
8FYE
 
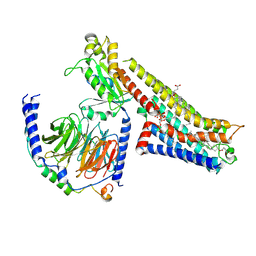 | | 4-F, 5-MeO-PyrT-bound serotonin 1A (5-HT1A) receptor-Gi1 protein complex | | Descriptor: | 4-fluoro-5-methoxy-3-[2-(pyrrolidin-1-yl)ethyl]-1H-indole, CHOLESTEROL HEMISUCCINATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Warren, A.L, Zilberg, G, Capper, M.J, Wacker, D. | | Deposit date: | 2023-01-26 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural pharmacology and therapeutic potential of 5-methoxytryptamines.
Nature, 630, 2024
|
|
6BGT
 
 | |
8GA6
 
 | |
8G9K
 
 | |
4WXR
 
 | |
4ZZY
 
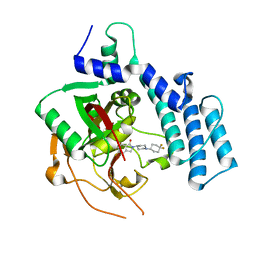 | | Structure of human PARP2 catalytic domain bound to an isoindolinone inhibitor | | Descriptor: | 2-[1-(4,4-Difluorocyclohexyl)-piperidin-4-yl]-6-fluoro-3-oxo-2,3-dihydro-1H-isoindole-4-carboxamide, POLY [ADP-RIBOSE] POLYMERASE 2 | | Authors: | Casale, E, Fasolini, M, Papeo, G, Posteri, H, Borghi, D, Busel, A.A, Caprera, F, Ciomei, M, Cirla, A, Corti, E, DAnello, M, Fasolini, M, Felder, E.R, Forte, B, Galvani, A, Isacchi, A, Khvat, A, Krasavin, M.Y, Lupi, R, Orsini, P, Perego, R, Pesenti, E, Pezzetta, D, Rainoldi, S, RiccardiSirtori, F, Scolaro, A, Sola, F, Zuccotto, F, Donati, D, Montagnoli, A. | | Deposit date: | 2015-04-15 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of 2-[1-(4,4-Difluorocyclohexyl)Piperidin-4-Yl]-6-Fluoro-3-Oxo-2,3-Dihydro-1H-Isoindole-4-Carboxamide (Nms-P118): A Potent, Orally Available and Highly Selective Parp- 1 Inhibitor for Cancer Therapy.
J.Med.Chem., 58, 2015
|
|
4ZZZ
 
 | | Structure of human PARP1 catalytic domain bound to an isoindolinone inhibitor | | Descriptor: | 2-(3-methoxypropyl)-3-oxo-2,3-dihydro-1H-isoindole-4-carboxamide, GLYCEROL, POLY [ADP-RIBOSE] POLYMERASE 1, ... | | Authors: | Casale, E, Fasolini, M, Papeo, G, Posteri, H, Borghi, D, Busel, A.A, Caprera, F, Ciomei, M, Cirla, A, Corti, E, DAnello, M, Fasolini, M, Felder, E.R, Forte, B, Galvani, A, Isacchi, A, Khvat, A, Krasavin, M.Y, Lupi, R, Orsini, P, Perego, R, Pesenti, E, Pezzetta, D, Rainoldi, S, RiccardiSirtori, F, Scolaro, A, Sola, F, Zuccotto, F, Donati, D, Montagnoli, A. | | Deposit date: | 2015-04-15 | | Release date: | 2015-08-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of 2-[1-(4,4-Difluorocyclohexyl)Piperidin-4-Yl]-6-Fluoro-3-Oxo-2,3-Dihydro-1H-Isoindole-4-Carboxamide (Nms-P118): A Potent, Orally Available and Highly Selective Parp- 1 Inhibitor for Cancer Therapy.
J.Med.Chem., 58, 2015
|
|
5A00
 
 | | Structure of human PARP1 catalytic domain bound to an isoindolinone inhibitor | | Descriptor: | 2-[1-(4,4-Difluorocyclohexyl)-piperidin-4-yl]-6-fluoro-3-oxo-2,3-dihydro-1H-isoindole-4-carboxamide, POLY [ADP-RIBOSE] POLYMERASE 1, SULFATE ION | | Authors: | Casale, E, Fasolini, M, Papeo, G, Posteri, H, Borghi, D, Busel, A.A, Caprera, F, Ciomei, M, Cirla, A, Corti, E, DAnello, M, Fasolini, M, Felder, E.R, Forte, B, Galvani, A, Isacchi, A, Khvat, A, Krasavin, M.Y, Lupi, R, Orsini, P, Perego, R, Pesenti, E, Pezzetta, D, Rainoldi, S, RiccardiSirtori, F, Scolaro, A, Sola, F, Zuccotto, F, Donati, D, Montagnoli, A. | | Deposit date: | 2015-04-15 | | Release date: | 2015-08-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Discovery of 2-[1-(4,4-Difluorocyclohexyl)Piperidin-4-Yl]-6-Fluoro-3-Oxo-2,3-Dihydro-1H-Isoindole-4-Carboxamide (Nms-P118): A Potent, Orally Available and Highly Selective Parp- 1 Inhibitor for Cancer Therapy.
J.Med.Chem., 58, 2015
|
|
2Y68
 
 | | Structure-based design of a new series of D-glutamic acid-based inhibitors of bacterial MurD ligase | | Descriptor: | 2-[[2-fluoro-5-[[[4-[(Z)-(4-oxo-2-sulfanylidene-1,3-thiazolidin-5-ylidene)methyl]phenyl]amino]methyl]phenyl]carbonylamino]pentanedioic acid, AZIDE ION, CHLORIDE ION, ... | | Authors: | Tomasic, T, Zidar, N, Sink, R, Kovac, A, Patin, D, Blanot, D, Contreras-Martel, C, Dessen, A, Muller-Premru, M, Zega, A, Gobec, S, Peterlin-Masic, L, Kikelj, D. | | Deposit date: | 2011-01-20 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structure-based design of a new series of D-glutamic acid based inhibitors of bacterial UDP-N-acetylmuramoyl-L-alanine:D-glutamate ligase (MurD).
J. Med. Chem., 54, 2011
|
|
6Y0Z
 
 | | X-ray structure of Lactobacillus brevis alcohol dehydrogenase mutant Q126K | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Hermann, J, Bischoff, D, Janowski, R, Niessing, D, Grob, P, Hekmat, D, Weuster-Botz, D. | | Deposit date: | 2020-02-10 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Controlling Protein Crystallization by Free Energy Guided Design of Interactions at Crystal Contacts
Crystals, 11, 2021
|
|
6Y10
 
 | | X-ray structure of Lactobacillus brevis alcohol dehydrogenase mutant Q126H | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, R-specific alcohol dehydrogenase | | Authors: | Hermann, J, Bischoff, D, Janowski, R, Niessing, D, Grob, P, Hekmat, D, Weuster-Botz, D. | | Deposit date: | 2020-02-10 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Controlling Protein Crystallization by Free Energy Guided Design of Interactions at Crystal Contacts
Crystals, 11, 2021
|
|
5ISN
 
 | | NMR solution structure of macro domain from Venezuelan equine encephalitis virus | | Descriptor: | Non-structural polyprotein | | Authors: | Makrynitsa, G.I, Ntonti, D, Marousis, K.D, Tsika, A.C, Papageorgiou, N, Coutard, B, Bentrop, D, Spyroulias, G.A. | | Deposit date: | 2016-03-15 | | Release date: | 2017-11-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Conformational plasticity of the VEEV macro domain is important for binding of ADP-ribose.
J.Struct.Biol., 206, 2019
|
|
6COA
 
 | | 1.2 A Structure of Thaumatin Crystallized in Gel | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Sauter, C, Lorber, B, Shabalin, I.G, Porebski, P.J, Brzezinski, D, Giege, R. | | Deposit date: | 2018-03-12 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Towards atomic resolution with crystals grown in gel: the case of thaumatin seen at room temperature.
Proteins, 48, 2002
|
|
6WXP
 
 | | De novo TIM barrel-ferredoxin fold fusion homodimer with 4-glutamate centre TFD-EE | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, TFD-EE | | Authors: | Caldwell, S.J, Zeymer, C, Haydon, I.C, Huang, P, Hilvert, D, Baker, D. | | Deposit date: | 2020-05-11 | | Release date: | 2020-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Tight and specific lanthanide binding in a de novo TIM barrel with a large internal cavity designed by symmetric domain fusion.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
