4Q2V
 
 | | Crystal Structure of Ricin A chain complexed with Baicalin inhibitor | | Descriptor: | 5,6-dihydroxy-4-oxo-2-phenyl-4H-chromen-7-yl beta-D-glucopyranosiduronic acid, Ricin | | Authors: | Deng, X, Li, X, Dong, J, Chen, Y. | | Deposit date: | 2014-04-10 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Baicalin inhibits the lethality of ricin in mice by inducing protein oligomerization.
J.Biol.Chem., 290, 2015
|
|
5EIG
 
 | |
5T18
 
 | | Crystal structure of Bruton agammabulinemia tyrosine kinase complexed with BMS-986142 aka (2s)-6-fluoro-5-[3-(8-fluoro-1-methyl-2,4-dioxo-1,2,3,4-tetrahydroquinazolin-3-yl)-2-methylphenyl]-2-(2-hydroxypropan-2-yl)-2,3,4,9-tetrahydro-1h-carbazole-8-carboxamide | | Descriptor: | 6-Fluoro-5-(R)-(3-(S)-(8-fluoro-1-methyl-2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)-2-methylphenyl)-2-(S)-(2-hydroxypropan-2-yl)-2,3,4,9-tetrahydro-1H-carbazole-8-carboxamide, Tyrosine-protein kinase BTK | | Authors: | Muckelbauer, J.K. | | Deposit date: | 2016-08-18 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of 6-Fluoro-5-(R)-(3-(S)-(8-fluoro-1-methyl-2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)-2-methylphenyl)-2-(S)-(2-hydroxypropan-2-yl)-2,3,4,9-tetrahydro-1H-carbazole-8-carboxamide (BMS-986142): A Reversible Inhibitor of Bruton's Tyrosine Kinase (BTK) Conformationally Constrained by Two Locked Atropisomers.
J. Med. Chem., 59, 2016
|
|
1YO6
 
 | |
4IN0
 
 | |
6OZ8
 
 | |
7DK8
 
 | | Crystal structure of OsGH3-8 with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Probable indole-3-acetic acid-amido synthetase GH3.8 | | Authors: | Zhang, Y.K, Xu, G.L, Ming, Z.H. | | Deposit date: | 2020-11-23 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structure of the acyl acid amido synthetase GH3-8 from Oryza sativa.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
5K86
 
 | |
6P1Y
 
 | | Crystal structure of Mtb aspartate decarboxylase mutant M117I | | Descriptor: | AMMONIUM ION, Aspartate 1-decarboxylase alpha chain, Aspartate 1-decarboxylase beta chain, ... | | Authors: | Sun, Q, Li, X, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2019-05-20 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | The molecular basis of pyrazinamide activity on Mycobacterium tuberculosis PanD.
Nat Commun, 11, 2020
|
|
6P9Y
 
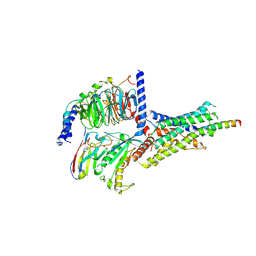 | | PAC1 GPCR Receptor complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Belousoff, M.J, Liang, Y.L, Sexton, P, Danev, R. | | Deposit date: | 2019-06-10 | | Release date: | 2020-02-05 | | Last modified: | 2020-02-19 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Toward a Structural Understanding of Class B GPCR Peptide Binding and Activation.
Mol.Cell, 77, 2020
|
|
6PNX
 
 | |
5KSJ
 
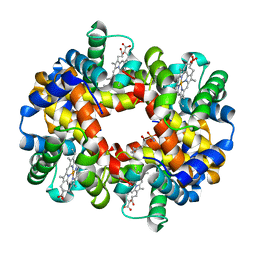 | | Crystal structure of deoxygenated hemoglobin in complex with Sphingosine phosphate | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Ahmed, M.H, Safo, M.K, Xia, Y. | | Deposit date: | 2016-07-08 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Functional Insight of Sphingosine 1-Phosphate-Mediated Pathogenic Metabolic Reprogramming in Sickle Cell Disease.
Sci Rep, 7, 2017
|
|
6PXG
 
 | |
6PXH
 
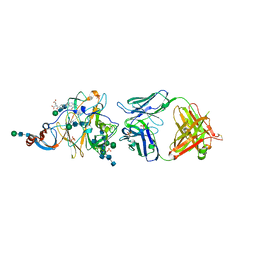 | | Crystal Structure of MERS-CoV S1-NTD bound with G2 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DIHYDROFOLIC ACID, ... | | Authors: | Wang, N, McLellan, J.S. | | Deposit date: | 2019-07-26 | | Release date: | 2019-09-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Definition of a Neutralization-Sensitive Epitope on the MERS-CoV S1-NTD.
Cell Rep, 28, 2019
|
|
1PPV
 
 | |
5KSI
 
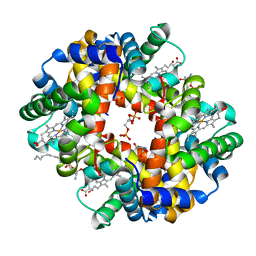 | | Crystal structure of deoxygenated hemoglobin in complex with sphingosine phosphate and 2,3-Bisphosphoglycerate | | Descriptor: | (2R)-2,3-diphosphoglyceric acid, (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Hemoglobin subunit alpha, ... | | Authors: | Ahmed, M.H, Safo, M.K, Xia, Y. | | Deposit date: | 2016-07-08 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Insight of Sphingosine 1-Phosphate-Mediated Pathogenic Metabolic Reprogramming in Sickle Cell Disease.
Sci Rep, 7, 2017
|
|
6PZ8
 
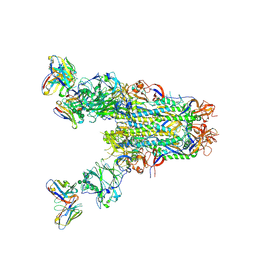 | | MERS S0 trimer in complex with variable domain of antibody G2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, G2 heavy chain, ... | | Authors: | Bowman, C.A, Pallesen, J, Ward, A.B. | | Deposit date: | 2019-07-31 | | Release date: | 2019-10-09 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.19 Å) | | Cite: | Structural Definition of a Neutralization-Sensitive Epitope on the MERS-CoV S1-NTD.
Cell Rep, 28, 2019
|
|
6P9X
 
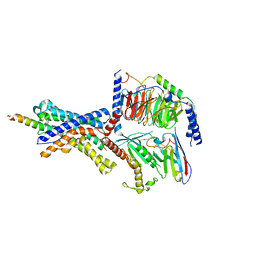 | | CRF1 Receptor Gs GPCR protein complex with CRF1 peptide | | Descriptor: | Corticoliberin, Corticotropin-releasing factor receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Belousoff, M.J, Liang, Y.L, Sexton, P, Danev, R. | | Deposit date: | 2019-06-10 | | Release date: | 2020-02-05 | | Last modified: | 2020-02-19 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Toward a Structural Understanding of Class B GPCR Peptide Binding and Activation.
Mol.Cell, 77, 2020
|
|
1PPW
 
 | |
6OYY
 
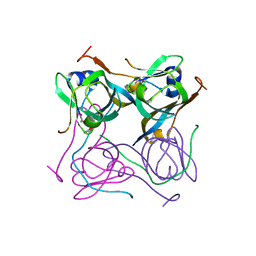 | | Crystal structure of Mtb aspartate decarboxylase, pyrazinoic acid complex | | Descriptor: | Aspartate 1-decarboxylase alpha chain, Aspartate 1-decarboxylase beta chain, PYRAZINE-2-CARBOXYLIC ACID | | Authors: | Sun, Q, Li, X, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2019-05-15 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The molecular basis of pyrazinamide activity on Mycobacterium tuberculosis PanD.
Nat Commun, 11, 2020
|
|
6P02
 
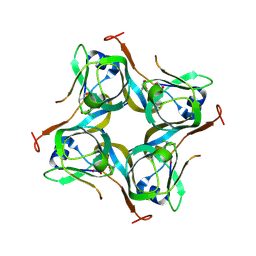 | | Crystal structure of Mtb aspartate decarboxylase, 6-Chlorine pyrazinoic acid complex | | Descriptor: | 6-chloropyrazine-2-carboxylic acid, Aspartate 1-decarboxylase alpha chain, Aspartate 1-decarboxylase beta chain | | Authors: | Sun, Q, Li, X, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2019-05-16 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The molecular basis of pyrazinamide activity on Mycobacterium tuberculosis PanD.
Nat Commun, 11, 2020
|
|
4Q05
 
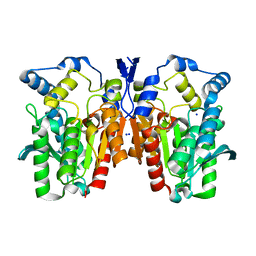 | | Crystal structure of an esterase E25 | | Descriptor: | SODIUM ION, esterase E25 | | Authors: | Li, P.Y, Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2014-03-31 | | Release date: | 2014-06-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for dimerization and catalysis of a novel esterase from the GTSAG motif subfamily of the bacterial hormone-sensitive lipase family
J.Biol.Chem., 289, 2014
|
|
4GA7
 
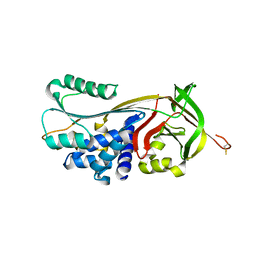 | | Crystal structure of human serpinB1 mutant | | Descriptor: | Leukocyte elastase inhibitor | | Authors: | Wang, L, Li, Q, Wu, L, Zhang, K, Tong, L, Sun, F, Fan, Z. | | Deposit date: | 2012-07-25 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification of SERPINB1 as a physiological inhibitor of human granzyme H
J.Immunol., 190, 2013
|
|
4GAW
 
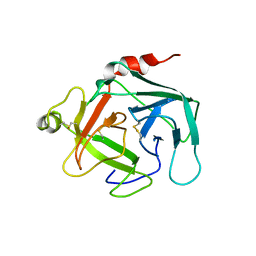 | | Crystal structure of active human granzyme H | | Descriptor: | CHLORIDE ION, Granzyme H, SULFATE ION | | Authors: | Wang, L, Li, Q, Wu, L, Zhang, K, Tong, L, Sun, F, Fan, Z. | | Deposit date: | 2012-07-25 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Identification of SERPINB1 as a physiological inhibitor of human granzyme H
J.Immunol., 190, 2013
|
|
7DFE
 
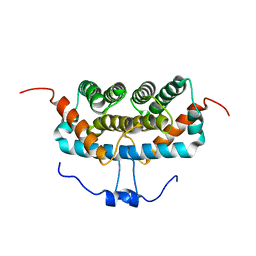 | | NMR structure of TuSp2-RP | | Descriptor: | B6 protein | | Authors: | Lin, Z, Fan, T, Fan, J. | | Deposit date: | 2020-11-07 | | Release date: | 2021-11-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1H, 15N and 13C resonance assignments of a repetitive domain of tubuliform spidroin 2
Biomol.Nmr Assign., 15, 2021
|
|
