8SVH
 
 | | Ubiquitin variant i53 mutant L67R bound to 53BP1 Tudor Domain | | Descriptor: | Tumor protein p53 binding protein 1, Ubiquitin variant i53: mutant L67R | | Authors: | Holden, J.K, Partridge, J.R, Wibowo, A.S, Mulichak, A. | | Deposit date: | 2023-05-16 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Functional screening in human HSPCs identifies optimized protein-based enhancers of Homology Directed Repair.
Nat Commun, 15, 2024
|
|
8SVI
 
 | | Ubiquitin variant i53:Mutant L67H with 53BP1 Tudor domain | | Descriptor: | GLYCEROL, Tumor protein p53 binding protein 1, Ubiquitin Variant i53: Mutant L67H | | Authors: | Partridge, J.R, Holden, J.K, Wibowo, A.S, Mulichak, A. | | Deposit date: | 2023-05-16 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Functional screening in human HSPCs identifies optimized protein-based enhancers of Homology Directed Repair.
Nat Commun, 15, 2024
|
|
6SE1
 
 | | Structure of Salmonella ser. Paratyphi A lipopolysaccharide acetyltransferase periplasmic domain | | Descriptor: | DI(HYDROXYETHYL)ETHER, Putative lipopolysaccharide modification acyltransferase, SULFATE ION | | Authors: | Tindall, S.N, Pearson, C, Herman, R, Jenkins, H.T, Thomas, G.H, Potts, J.R, Van der Woude, M. | | Deposit date: | 2019-07-29 | | Release date: | 2020-08-26 | | Last modified: | 2020-09-16 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Acetylation of Surface Carbohydrates in Bacterial Pathogens Requires Coordinated Action of a Two-Domain Membrane-Bound Acyltransferase.
Mbio, 11, 2020
|
|
8SVJ
 
 | | Ubiquitin variant i53: mutant VHH with 53BP1 Tudor domain | | Descriptor: | GLYCEROL, Tumor protein p53 binding protein 1, Ubiquitin varient i53 mutant VHH | | Authors: | Holden, J, Partridge, J.R, Wibowo, A.S, Mulichak, A. | | Deposit date: | 2023-05-16 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Functional screening in human HSPCs identifies optimized protein-based enhancers of Homology Directed Repair.
Nat Commun, 15, 2024
|
|
8T2D
 
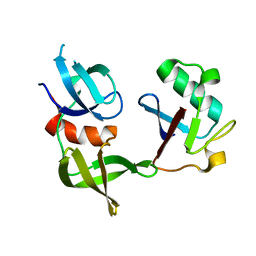 | | Ubiquitin variant i53:Mutant T12Y.T14E.L67R with 53BP1 Tudor domain | | Descriptor: | Tumor protein p53 binding protein 1, Ubiquitin variant i53 | | Authors: | Partridge, J.R, Holden, J.K, Wibowo, A.S, Mulichak, A. | | Deposit date: | 2023-06-05 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Functional screening in human HSPCs identifies optimized protein-based enhancers of Homology Directed Repair.
Nat Commun, 15, 2024
|
|
8T0O
 
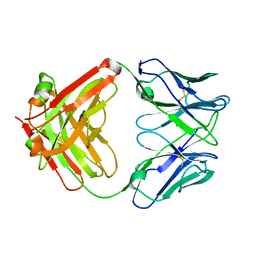 | | Fab from mAb RB2AT_87 | | Descriptor: | CHLORIDE ION, RB2AT_87 Fab Heavy chain, RB2AT_87 Fab Light chain | | Authors: | Kreutzer, A.G, Malonis, R.J, Lai, J.R, Nowick, J.S. | | Deposit date: | 2023-06-01 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Generation and Study of Antibodies against Two Triangular Trimers Derived from A beta.
Pept Sci (Hoboken), 116, 2024
|
|
2LV2
 
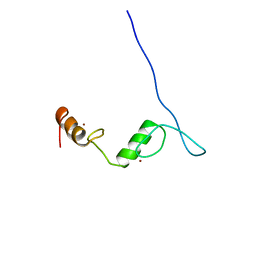 | | Solution NMR structure of C2H2-type Zinc-fingers 4 and 5 from human Insulinoma-associated protein 1 (fragment 424-497), Northeast Structural Genomics Consortium Target HR7614B | | Descriptor: | Insulinoma-associated protein 1, ZINC ION | | Authors: | Yang, Y, Ramelot, T.A, Cort, J.R, Shastry, R, Kohan, E, Janjua, H, Xiao, R, Acton, T, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-06-27 | | Release date: | 2012-10-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of C2H2-type Zinc-fingers 4 and 5 from human Insulinoma-associated protein 1 (fragment 424-497), Northeast Structural
Genomics Consortium Target HR7614B (CASP Target)
To be Published
|
|
1SNO
 
 | |
2M4H
 
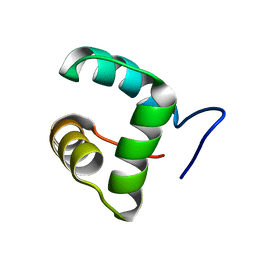 | | Solution structure of the Core Domain (10-76) of the Feline Calicivirus VPg protein | | Descriptor: | Feline Calicivirus VPg protein | | Authors: | Kwok, R.N, Leen, E.N, Birtley, J.R, Prater, S.N, Simpson, P.J, Curry, S, Matthews, S, Marchant, J. | | Deposit date: | 2013-02-05 | | Release date: | 2013-03-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structures of the Compact Helical Core Domains of Feline Calicivirus and Murine Norovirus VPg Proteins.
J.Virol., 87, 2013
|
|
8SZ8
 
 | |
8SZ7
 
 | |
8SZ4
 
 | |
8T0K
 
 | |
8SXZ
 
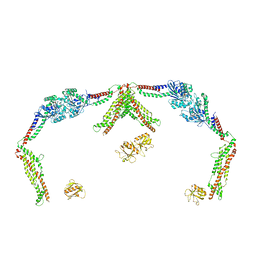 | |
8T0R
 
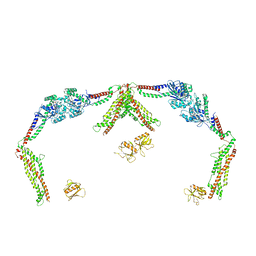 | |
2MA5
 
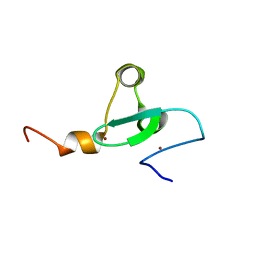 | | Solution NMR structure of PHD type Zinc finger domain of Lysine-specific demethylase 5B (PLU-1/JARID1B) from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR7375C | | Descriptor: | Lysine-specific demethylase 5B, ZINC ION | | Authors: | Hassan, F, Ramelot, T.A, Yang, Y, Cort, J.R, Janjua, H, Kohan, E, Lee, D, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-06-28 | | Release date: | 2013-08-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of PHD type Zinc finger domain of Lysine-specific
demethylase 5B (PLU-1/JARID1B) from Homo sapiens, Northeast Structural
Genomics Consortium (NESG) Target HR7375C
To be Published
|
|
1TJG
 
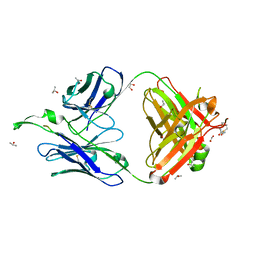 | | Crystal Structure of the broadly neutralizing anti-HIV-1 antibody 2F5 in complex with a gp41 7mer epitope | | Descriptor: | 1,2-ETHANEDIOL, Envelope glycoprotein GP41, FAB 2F5 Heavy Chain, ... | | Authors: | Ofek, G, Tang, M, Sambor, A, Katinger, H, Mascola, J.R, Wyatt, R, Kwong, P.D. | | Deposit date: | 2004-06-04 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and mechanistic analysis of the Anti-Human Immunodeficiency Virus Type 1 antibody 2F5 in complex with its gp41 epitope
J.Virol., 78, 2004
|
|
3MZL
 
 | | Sec13/Sec31 edge element, loop deletion mutant | | Descriptor: | Protein transport protein SEC13, Protein transport protein SEC31 | | Authors: | Whittle, J.R, Schwartz, T.U. | | Deposit date: | 2010-05-12 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Sec13-Sec16 edge element, a template for assembly of the COPII vesicle coat.
J.Cell Biol., 190, 2010
|
|
1UD7
 
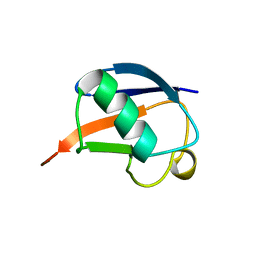 | | SOLUTION STRUCTURE OF THE DESIGNED HYDROPHOBIC CORE MUTANT OF UBIQUITIN, 1D7 | | Descriptor: | PROTEIN (UBIQUITIN CORE MUTANT 1D7) | | Authors: | Johnson, E.C, Lazar, G.A, Desjarlais, J.R, Handel, T.M. | | Deposit date: | 1999-04-07 | | Release date: | 1999-05-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of a designed hydrophobic core variant of ubiquitin.
Structure Fold.Des., 7, 1999
|
|
8SOZ
 
 | |
8SOW
 
 | |
3NGG
 
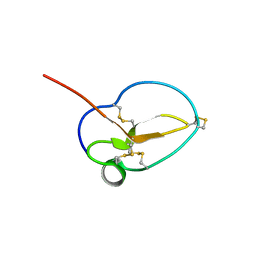 | | X-ray Structure of Omwaprin | | Descriptor: | Omwaprin-a | | Authors: | Banigan, J.R, Mandal, K, Sawaya, M.R, Thammavongsa, V, Hendrickx, A.P.A, Schneewind, O, Yeates, T.O, Kent, S.B.H. | | Deposit date: | 2010-06-11 | | Release date: | 2010-09-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Determination of the X-ray structure of the snake venom protein omwaprin by total chemical synthesis and racemic protein crystallography.
Protein Sci., 19, 2010
|
|
1U35
 
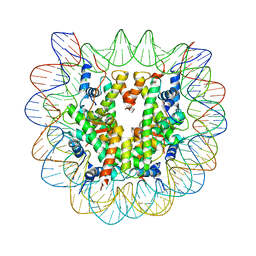 | | Crystal structure of the nucleosome core particle containing the histone domain of macroH2A | | Descriptor: | H2A histone family, Hist1h4i protein, Histone H3.1, ... | | Authors: | Chakravarthy, S, Gundimella, S.K, Caron, C, Perche, P.Y, Pehrson, J.R, Khochbin, S, Luger, K. | | Deposit date: | 2004-07-20 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural characterization of the histone variant macroH2A.
Mol.Cell.Biol., 25, 2005
|
|
8U4U
 
 | |
1U00
 
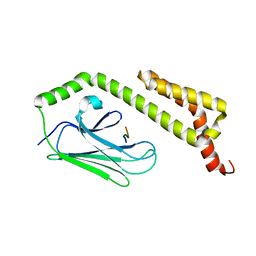 | | HscA substrate binding domain complexed with the IscU recognition peptide ELPPVKIHC | | Descriptor: | Chaperone protein hscA, IscU recognition peptide | | Authors: | Cupp-Vickery, J.R, Peterson, J.C, Ta, D.T, Vickery, L.E. | | Deposit date: | 2004-07-12 | | Release date: | 2004-10-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of the Molecular Chaperone HscA Substrate Binding Domain Complexed with the IscU Recognition Peptide ELPPVKIHC.
J.Mol.Biol., 342, 2004
|
|
