4V39
 
 | | Apo-structure of alpha2,3-sialyltransferase variant 2 from Pasteurella dagmatis | | Descriptor: | SIALYLTRANSFERASE | | Authors: | Pavkov-Keller, T, Schmoelzer, K, Czabany, T, Luley-Goedl, C, Ribitsch, D, Schwab, H, Nidetzky, B, Gruber, K. | | Deposit date: | 2014-10-17 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Complete Switch from Alpha2,3- to Alpha2,6-Regioselectivity in Pasteurella Dagmatis Beta-D-Galactoside Sialyltransferase by Active-Site Redesign
Chem.Commun.(Camb.), 51, 2015
|
|
4UWK
 
 | | Discovery of (2S)-8-((3R)-3-Methylmorpholin-4-yl)-1-(3-methyl-2-oxo- butyl)-2-(trifluoromethyl)-3,4-dihydro-2H-pyrimido(1,2-a)pyrimidin-6- one: a Novel Potent and Selective Inhibitor of Vps34 for the Treatment of Solid Tumors | | Descriptor: | (2S)-1-[(5-chloro-2-thienyl)methyl]-8-[(3R,5R)-3,5-dimethylmorpholin-4-yl]-2-(trifluoromethyl)-3,4-dihydro-2H-pyrimido[1,2-a]pyrimidin-6-one, GLYCEROL, PHOSPHATIDYLINOSITOL 3-KINASE CATALYTIC SUBUNIT TYPE 3, ... | | Authors: | Pasquier, B, El-Ahmad, Y, Filoche-Romme, B, Dureuil, C, Fassy, F, Abecassis, P.Y, Mathieu, M, Bertrand, T, Benard, T, Barriere, C, ElBatti, S, Letallec, J.P, Sonnefraud, V, Brollo, M, Delbarre, L, Loyau, V, Pilorge, F, Bertin, L, Richepin, P, Arigon, J, Labrosse, J.R, Clement, J, Durand, F, Combet, R, Perraut, P, Leroy, V, Gay, F, Lefrancois, D, Bretin, F, Marquette, J.P, Michot, N, Caron, A, Castell, C, Schio, L, McCort, G, Goulaouic, H, Garcia-Echeverria, C, Ronan, B. | | Deposit date: | 2014-08-12 | | Release date: | 2014-11-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Discovery of (2S)-8-[(3R)-3-Methylmorpholin-4-Yl]-1-(3-Methyl-2-Oxo-Butyl)-2-(Trifluoromethyl)-3,4-Dihydro-2H-Pyrimido[1,2-A]Pyrimidin-6-One: A Novel Potent and Selective Inhibitor of Vps34 for the Treatment of Solid Tumors.
J.Med.Chem., 58, 2015
|
|
4UWL
 
 | | Discovery of (2S)-8-((3R)-3-Methylmorpholin-4-yl)-1-(3-methyl-2-oxo- butyl)-2-(trifluoromethyl)-3,4-dihydro-2H-pyrimido(1,2-a)pyrimidin-6- one: a Novel Potent and Selective Inhibitor of Vps34 for the Treatment of Solid Tumors | | Descriptor: | (8S)-2-[(3R)-3-methylmorpholin-4-yl]-9-(3-methyl-2-oxobutyl)-8-(trifluoromethyl)-6,7,8,9-tetrahydro-4H-pyrimido[1,2-a]pyrimidin-4-one, PHOSPHATIDYLINOSITOL 3-KINASE CATALYTIC SUBUNIT TYPE 3, SULFATE ION | | Authors: | Pasquier, B, El-Ahmad, Y, Filoche-Romme, B, Dureuil, C, Fassy, F, Abecassis, P.Y, Mathieu, M, Bertrand, T, Benard, T, Barriere, C, ElBatti, S, Letallec, J.P, Sonnefraud, V, Brollo, M, Delbarre, L, Loyau, V, Pilorge, F, Bertin, L, Richepin, P, Arigon, J, Labrosse, J.R, Clement, J, Durand, F, Combet, R, Perraut, P, Leroy, V, Gay, F, Lefrancois, D, Bretin, F, Marquette, J.P, Michot, N, Caron, A, Castell, C, Schio, L, McCort, G, Goulaouic, H, Garcia-Echeverria, C, Ronan, B. | | Deposit date: | 2014-08-12 | | Release date: | 2014-11-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of (2S)-8-[(3R)-3-Methylmorpholin-4-Yl]-1-(3-Methyl-2-Oxo-Butyl)-2-(Trifluoromethyl)-3,4-Dihydro-2H-Pyrimido[1,2-A]Pyrimidin-6-One: A Novel Potent and Selective Inhibitor of Vps34 for the Treatment of Solid Tumors.
J.Med.Chem., 58, 2015
|
|
4V0W
 
 | | The crystal structure of mouse PP1G in complex with truncated human PPP1R15B (631-669) | | Descriptor: | MANGANESE (II) ION, PROTEIN PHOSPHATASE 1 REGULATORY SUBUNIT 15B, SERINE/THREONINE-PROTEIN PHOSPHATASE PP1-GAMMA CATALYTIC SUBUNIT | | Authors: | Chen, R, Yan, Y, Casado, A.C, Ron, D, Read, R.J. | | Deposit date: | 2014-09-18 | | Release date: | 2015-03-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | G-actin provides substrate-specificity to eukaryotic initiation factor 2 alpha holophosphatases.
Elife, 4, 2015
|
|
3VBH
 
 | | Crystal structure of formaldehyde treated human enterovirus 71 (space group R32) | | Descriptor: | CHLORIDE ION, Genome Polyprotein, capsid protein VP1, ... | | Authors: | Wang, X, Peng, W, Ren, J, Hu, Z, Xu, J, Lou, Z, Li, X, Yin, W, Shen, X, Porta, C, Walter, T.S, Evans, G, Axford, D, Owen, R, Rowlands, D.J, Wang, J, Stuart, D.I, Fry, E.E, Rao, Z. | | Deposit date: | 2012-01-02 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3VCK
 
 | | Thaumatin by LB Hanging Drop Vapour Diffusion after 9.05 MGy X-Ray dose at ESRF ID29 beamline (Worst Case) | | Descriptor: | GLYCEROL, Thaumatin I | | Authors: | Belmonte, L, Pechkova, E, Scudieri, D, Nicolini, C. | | Deposit date: | 2012-01-04 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Langmuir-blodgett nanotemplate and radiation resistance in protein crystals: state of the art.
Crit Rev Eukaryot Gene Expr, 22, 2012
|
|
3VDM
 
 | | Crystal Structure of VldE, the pseudo-glycosyltransferase which catalyzes non-glycosidic C-N coupling in Validamycin A biosynthesis | | Descriptor: | VldE | | Authors: | Cavalier, M.C, Yim, Y.-S, Asamizu, S, Neau, D, Mahmud, T, Lee, Y.-H. | | Deposit date: | 2012-01-05 | | Release date: | 2013-01-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal Structure of VldE, the pseudo-glycosyltransferase which catalyzes non-glycosidic C-N coupling in Validamycin A biosynthesis
To be Published
|
|
4UX5
 
 | | Structure of DNA complex of PCG2 | | Descriptor: | 5'-D(*CP*AP*AP*TP*GP*AP*CP*GP*CP*GP*TP*AP*AP*GP)-3', 5'-D(*CP*TP*TP*AP*CP*GP*CP*GP*TP*CP*AP*TP*TP*GP)-3', TRANSCRIPTION FACTOR MBP1 | | Authors: | Liu, J, Huang, J, Zhao, Y, Liu, H, Wang, D, Yang, J, Zhao, W, Taylor, I.A, Peng, Y. | | Deposit date: | 2014-08-19 | | Release date: | 2015-01-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of DNA Recognition by Pcg2 Reveals a Novel DNA Binding Mode for Winged Helix-Turn-Helix Domains.
Nucleic Acids Res., 43, 2015
|
|
4UXH
 
 | | Leishmania major Thymidine Kinase in complex with AP5dT | | Descriptor: | P1-(5'-ADENOSYL)P5-(5'-THYMIDYL)PENTAPHOSPHATE, THYMIDINE KINASE, ZINC ION | | Authors: | Timm, J, Bosch-Navarrete, C, Recio, E, Nettleship, J.E, Rada, H, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2014-08-22 | | Release date: | 2015-05-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Kinetic Characterization of Thymidine Kinase from Leishmania Major.
Plos Negl Trop Dis, 9, 2015
|
|
4V1X
 
 | | The structure of the hexameric atrazine chlorohydrolase, AtzA | | Descriptor: | ATRAZINE CHLOROHYDROLASE, DI(HYDROXYETHYL)ETHER, FE (III) ION | | Authors: | Peat, T.S, Newman, J, Balotra, S, Lucent, D, Warden, A.C, Scott, C. | | Deposit date: | 2014-10-04 | | Release date: | 2015-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of the Hexameric Atrazine Chlorohydrolase Atza.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UYI
 
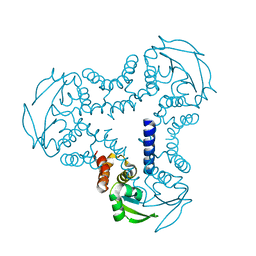 | | Crystal structure of the BTB domain of human SLX4 (BTBD12) | | Descriptor: | STRUCTURE-SPECIFIC ENDONUCLEASE SUBUNIT SLX4 | | Authors: | Pinkas, D.M, Sanvitale, C.E, Strain-Damerell, C, Fairhead, M, Wang, D, Tallant, C, Cooper, C.D.O, Sorrell, F.J, Kopec, J, Chaikuad, A, Fitzpatrick, F, Pike, A.C.W, Hozjan, V, Ying, Z, Roos, A.K, Savitsky, P, Bradley, A, Nowak, R, Filippakopoulos, P, Krojer, T, Burgess-Brown, N.A, Marsden, B.D, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2014-09-01 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal Structure of the Btb Domain of Human Slx4 (Btbd12)
To be Published
|
|
4UXJ
 
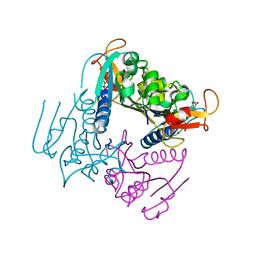 | | Leishmania major Thymidine Kinase in complex with dTTP | | Descriptor: | MAGNESIUM ION, THYMIDINE KINASE, THYMIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Timm, J, Bosch-Navarrete, C, Recio, E, Nettleship, J.E, Rada, H, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2014-08-22 | | Release date: | 2015-05-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Kinetic Characterization of Thymidine Kinase from Leishmania Major.
Plos Negl Trop Dis, 9, 2015
|
|
3VZ2
 
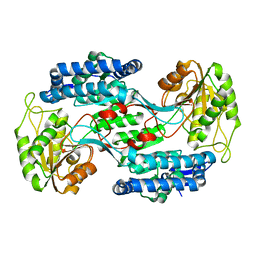 | |
4UXI
 
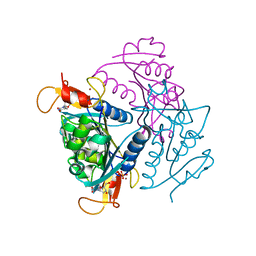 | | Leishmania major Thymidine Kinase in complex with thymidine | | Descriptor: | PHOSPHATE ION, THYMIDINE, THYMIDINE KINASE, ... | | Authors: | Timm, J, Bosch-Navarrete, C, Recio, E, Nettleship, J.E, Rada, H, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2014-08-22 | | Release date: | 2015-05-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural and Kinetic Characterization of Thymidine Kinase from Leishmania Major.
Plos Negl Trop Dis, 9, 2015
|
|
3VZ1
 
 | |
3V82
 
 | | Thaumatin by LB based Hanging Drop Vapour Diffusion after 1.81 MGy X-Ray dose at ESRF ID29 beamline (Best Case) | | Descriptor: | GLYCEROL, Thaumatin I | | Authors: | Belmonte, L, Scudieri, D, Tripathi, S, Pechkova, E, Nicolini, C. | | Deposit date: | 2011-12-22 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Langmuir-Blodgett nanotemplate and radiation resistance in protein crystals: state of the art.
CRIT.REV.EUKARYOT.GENE EXPR., 22, 2012
|
|
3V8C
 
 | | Crystal structure of monoclonal human anti-rhesus D Fc IgG1 t125(yb2/0) double mutant (H310 and H435 in K) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose, GLYCEROL, ... | | Authors: | Menez, R, Stura, E.A, Bourel, D, Siberil, S, Jorieux, S, De Romeuf, C, Ducancel, F, Fridman, W.H, Teillaud, J.L. | | Deposit date: | 2011-12-22 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Effect of zinc on human IgG1 and its Fc gamma R interactions.
Immunol.Lett., 143, 2012
|
|
3V95
 
 | | Crystal structure of monoclonal human anti-rhesus D Fc and IgG1 t125(yb2/0) in the presence of EDTA | | Descriptor: | GLYCEROL, Ig gamma-1 chain C region, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Menez, R, A Stura, E, Bourel, D, Siberil, S, Jorieux, S, De Romeuf, C, Ducancel, F, Fridman, W.H, Teillaud, J.L. | | Deposit date: | 2011-12-23 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Effect of zinc on human IgG1 and its Fc gamma R interactions.
Immunol.Lett., 143, 2012
|
|
4WH7
 
 | | Structure of the CDC25B Phosphatase Catalytic Domain with Bound Ligand | | Descriptor: | 2-fluoro-4-hydroxybenzonitrile, GLYCEROL, M-phase inducer phosphatase 2, ... | | Authors: | Lund, G.L, Dudkin, S, Borkin, D, Ni, W, Grembecka, J, Cierpicki, T. | | Deposit date: | 2014-09-20 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Inhibition of CDC25B Phosphatase Through Disruption of Protein-Protein Interaction.
Acs Chem.Biol., 10, 2015
|
|
3VCI
 
 | | Thaumatin by Classical Hanging Drop Vapour Diffusion after 9.05 MGy X-Ray dose at ESRF ID29 beamline (Worst Case) | | Descriptor: | GLYCEROL, Thaumatin I | | Authors: | Belmonte, L, Pechkova, E, Scudieri, D, Nicolini, C. | | Deposit date: | 2012-01-04 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Langmuir-blodgett nanotemplate and radiation resistance in protein crystals: state of the art.
Crit Rev Eukaryot Gene Expr, 22, 2012
|
|
4WH9
 
 | | Structure of the CDC25B Phosphatase Catalytic Domain with Bound Inhibitor | | Descriptor: | 2-[(2-cyano-3-fluoro-5-hydroxyphenyl)sulfanyl]ethanesulfonic acid, GLYCEROL, M-phase inducer phosphatase 2, ... | | Authors: | Lund, G.L, Dudkin, S, Borkin, D, Ni, W, Grembecka, J, Cierpicki, T. | | Deposit date: | 2014-09-20 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Inhibition of CDC25B Phosphatase Through Disruption of Protein-Protein Interaction.
Acs Chem.Biol., 10, 2015
|
|
3VNC
 
 | | Crystal Structure of TIP-alpha N25 from Helicobacter Pylori in its natural dimeric form | | Descriptor: | TIP-alpha | | Authors: | Gao, M, Li, D, Hu, Y, Zou, Q, Wang, D.-C. | | Deposit date: | 2012-01-11 | | Release date: | 2012-10-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of TNF-alpha-Inducing Protein from Helicobacter Pylori in Active Form Reveals the Intrinsic Molecular Flexibility for Unique DNA-Binding
Plos One, 7, 2012
|
|
3VQF
 
 | | Crystal Structure Analysis of the PDZ Domain Derived from the Tight Junction Regulating Protein | | Descriptor: | E3 ubiquitin-protein ligase LNX | | Authors: | Akiyoshi, Y, Hamada, D, Goda, N, Tenno, T, Narita, H, Nakagawa, A, Furuse, M, Suzuki, M, Hiroaki, H. | | Deposit date: | 2012-03-22 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | Structural basis for down regulation of tight junction by PDZ-domain containing E3-Ubiquitin ligase
To be Published
|
|
4WHX
 
 | | X-ray Crystal Structure of an Amino Acid Aminotransferase from Burkholderia pseudomallei Bound to the Co-factor Pyridoxal Phosphate | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, Branched-chain-amino-acid transaminase | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID), Fairman, J.W, Dranow, D.M, Taylor, B.M, Lorimer, D, Edwards, T.E. | | Deposit date: | 2014-09-23 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray Crystal Structure of an Amino Acid Aminotransferase from Burkholderia pseudomallei Bound to the Co-factor Pyridoxal Phosphate
to be published
|
|
3VJI
 
 | | Human PPAR gamma ligand binding domain in complex with JKPL53 | | Descriptor: | (2S)-2-{4-butoxy-3-[({4-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]benzoyl}amino)methyl]benzyl}butanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Tomioka, D, Kuwabara, N, Hashimoto, H, Sato, M, Shimizu, T. | | Deposit date: | 2011-10-20 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Peroxisome proliferator-activated receptors (PPARs) have multiple binding points that accommodate ligands in various conformations: phenylpropanoic acid-type PPAR ligands bind to PPAR in different conformations, depending on the subtype.
J.Med.Chem., 55, 2012
|
|
