7MEA
 
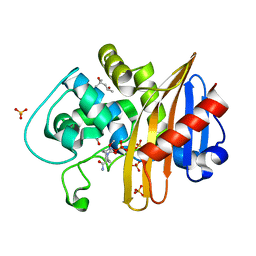 | | CDD-1 beta-lactamase in imidazole/MPD 1 minute avibactam complex | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-lactamase, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2021-04-06 | | Release date: | 2022-02-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | In Crystallo Time-Resolved Interaction of the Clostridioides difficile CDD-1 enzyme with Avibactam Provides New Insights into the Catalytic Mechanism of Class D beta-lactamases.
Acs Infect Dis., 7, 2021
|
|
7MED
 
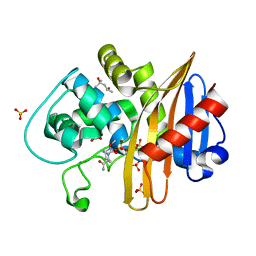 | | CDD-1 beta-lactamase in imidazole/MPD 5 minute avibactam complex | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-lactamase, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2021-04-06 | | Release date: | 2022-02-16 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | In Crystallo Time-Resolved Interaction of the Clostridioides difficile CDD-1 enzyme with Avibactam Provides New Insights into the Catalytic Mechanism of Class D beta-lactamases.
Acs Infect Dis., 7, 2021
|
|
7MEB
 
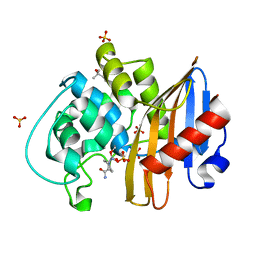 | | CDD-1 beta-lactamase in imidazole/MPD 2 minute avibactam complex | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-lactamase, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2021-04-06 | | Release date: | 2022-02-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In Crystallo Time-Resolved Interaction of the Clostridioides difficile CDD-1 enzyme with Avibactam Provides New Insights into the Catalytic Mechanism of Class D beta-lactamases.
Acs Infect Dis., 7, 2021
|
|
7MEH
 
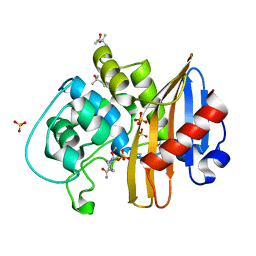 | | CDD-1 beta-lactamase in imidazole/MPD 60 minute avibactam complex | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-lactamase, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2021-04-06 | | Release date: | 2022-02-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In Crystallo Time-Resolved Interaction of the Clostridioides difficile CDD-1 enzyme with Avibactam Provides New Insights into the Catalytic Mechanism of Class D beta-lactamases.
Acs Infect Dis., 7, 2021
|
|
7LNR
 
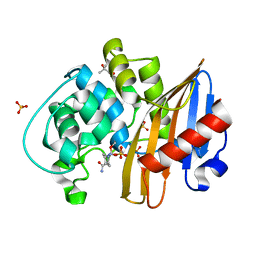 | | Structure of the avibactam-CDD-1 120 minute complex in imidazole and MPD | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-lactamase, ... | | Authors: | Smith, C.A, Vakulenko, S.B, Stewart, N.K. | | Deposit date: | 2021-02-08 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | In Crystallo Time-Resolved Interaction of the Clostridioides difficile CDD-1 enzyme with Avibactam Provides New Insights into the Catalytic Mechanism of Class D beta-lactamases.
Acs Infect Dis., 7, 2021
|
|
7LNO
 
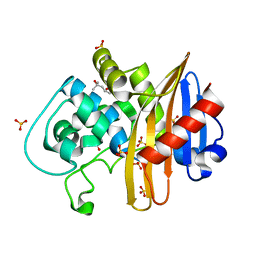 | | Structure of apo-CDD-1 beta-lactamase in imidazole and MPD | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-lactamase, SULFATE ION | | Authors: | Smith, C.A, Vakulenko, S.B, Stewart, N.K. | | Deposit date: | 2021-02-08 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | In Crystallo Time-Resolved Interaction of the Clostridioides difficile CDD-1 enzyme with Avibactam Provides New Insights into the Catalytic Mechanism of Class D beta-lactamases.
Acs Infect Dis., 7, 2021
|
|
7LNQ
 
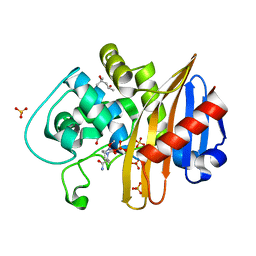 | | Structure of the avibactam-CDD-1 3 minute complex in imidazole and MPD | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-lactamase, ... | | Authors: | Smith, C.A, Vakulenko, S.B, Stewart, N.K. | | Deposit date: | 2021-02-08 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | In Crystallo Time-Resolved Interaction of the Clostridioides difficile CDD-1 enzyme with Avibactam Provides New Insights into the Catalytic Mechanism of Class D beta-lactamases.
Acs Infect Dis., 7, 2021
|
|
3N4U
 
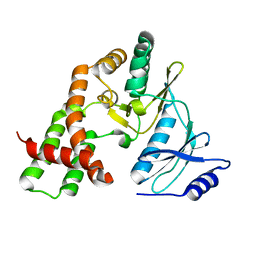 | |
3N4V
 
 | |
3N4T
 
 | | apo APH(2")-IVa form I | | Descriptor: | APH(2'')-Id | | Authors: | Smith, C.A, Toth, M, Frase, H, Vakulenko, S.B. | | Deposit date: | 2010-05-22 | | Release date: | 2010-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and kinetic mechanism of aminoglycoside phosphotransferase-2''-IVa.
Protein Sci., 19, 2010
|
|
1ND4
 
 | | Crystal structure of aminoglycoside-3'-phosphotransferase-IIa | | Descriptor: | ACETATE ION, Aminoglycoside 3'-phosphotransferase, KANAMYCIN A, ... | | Authors: | Nurizzo, D, Shewry, S.C, Baker, E.N, Smith, C.A. | | Deposit date: | 2002-12-06 | | Release date: | 2003-09-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of aminoglycoside-3'-phosphotransferase-IIa, an enzyme responsible for antibiotic resistance
J.Mol.Biol., 327, 2003
|
|
4QU3
 
 | | GES-2 ertapenem acyl-enzyme complex | | Descriptor: | (4R,5S)-3-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase GES-2, ... | | Authors: | Stewart, N.K, Smith, C.A, Frase, H, Black, D.J, Vakulenko, S.B. | | Deposit date: | 2014-07-10 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.402 Å) | | Cite: | Kinetic and Structural Requirements for Carbapenemase Activity in GES-Type beta-Lactamases.
Biochemistry, 54, 2015
|
|
4REV
 
 | | Structure of the dirigent protein DRR206 | | Descriptor: | 2,3-DIMETHYLIMIDAZOLIUM ION, CHLORIDE ION, Disease resistance response protein 206, ... | | Authors: | Kim, K.-Y, Smith, C.A, Merkley, E.D, Cort, J.R, Davin, L.B, Lewis, N.G. | | Deposit date: | 2014-09-24 | | Release date: | 2014-11-26 | | Last modified: | 2015-02-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Trimeric Structure of (+)-Pinoresinol-forming Dirigent Protein at 1.95 angstrom Resolution with Three Isolated Active Sites.
J.Biol.Chem., 290, 2015
|
|
8DA2
 
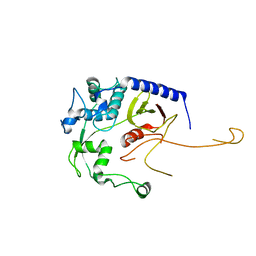 | | Acinetobacter baumannii L,D-transpeptidase | | Descriptor: | L,D-transpeptidase family protein | | Authors: | Toth, M, Stewart, N.K, Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2022-06-12 | | Release date: | 2022-09-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The l,d-Transpeptidase Ldt Ab from Acinetobacter baumannii Is Poorly Inhibited by Carbapenems and Has a Unique Structural Architecture.
Acs Infect Dis., 8, 2022
|
|
4NET
 
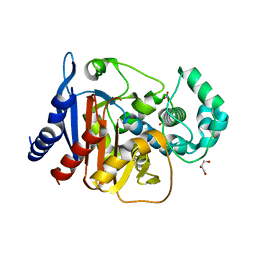 | | Crystal structure of ADC-1 beta-lactamase | | Descriptor: | AmpC, GLYCEROL, NITRATE ION | | Authors: | Bhattacharya, M, Toth, M, Antunes, N.T, Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2013-10-30 | | Release date: | 2014-03-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of the extended-spectrum class C beta-lactamase ADC-1 from Acinetobacter baumannii.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4N5H
 
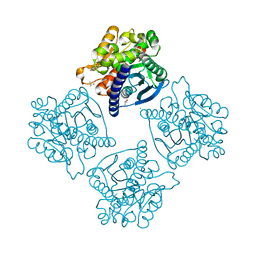 | | Crystal structure of ESTERASE B from Lactobacillus Rhamnosis (HN001) | | Descriptor: | CALCIUM ION, CHLORIDE ION, Esterase/lipase, ... | | Authors: | Bennett, M.D, Holland, R, Loo, T.S, Smith, C.A, Norris, G.E, Delabre, M.-L, Anderson, B.F. | | Deposit date: | 2013-10-09 | | Release date: | 2014-10-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal structure of ESTERASE B from Lactobacillus Rhamnosis (HN001)
TO BE PUBLISHED
|
|
2F00
 
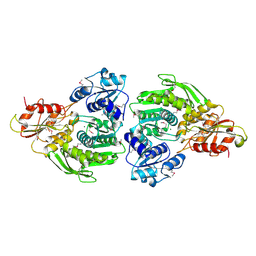 | | Escherichia coli MurC | | Descriptor: | MAGNESIUM ION, UDP-N-acetylmuramate--L-alanine ligase | | Authors: | Deva, T, Baker, E.N, Squire, C.J, Smith, C.A. | | Deposit date: | 2005-11-10 | | Release date: | 2006-10-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Escherichia coliUDP-N-acetylmuramoyl:L-alanine ligase (MurC).
Acta Crystallogr.,Sect.D, 62, 2006
|
|
6OTW
 
 | | Crystallographic Structure of (HbII-HbIII)-O2 from Lucina pectinata at pH 5.0 | | Descriptor: | Hemoglobin II, Hemoglobin III, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Marchany-Rivera, D, Smith, C.A, Rodriguez-Perez, J.D, Lopez-Garriga, J. | | Deposit date: | 2019-05-03 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.447 Å) | | Cite: | Lucina pectinata oxyhemoglobin (II-III) heterodimer pH susceptibility.
J.Inorg.Biochem., 207, 2020
|
|
6OTY
 
 | | Crystallographic Structure of (HbII-HbIII)-O2 from Lucina pectinata at pH 4.0 | | Descriptor: | Hemoglobin II, Hemoglobin III, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Marchany-Rivera, D, Smith, C.A, Rodriguez-Perez, J.D, Lopez-Garriga, J. | | Deposit date: | 2019-05-03 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Lucina pectinata oxyhemoglobin (II-III) heterodimer pH susceptibility.
J.Inorg.Biochem., 207, 2020
|
|
6OTX
 
 | | Crystallographic Structure of (HbII-HbIII)-O2 from Lucina pectinata at pH 7.0 | | Descriptor: | Hemoglobin II, Hemoglobin III, OXYGEN MOLECULE, ... | | Authors: | Marchany-Rivera, D, Smith, C.A, Rodriguez-Perez, J.D, Lopez-Garriga, J. | | Deposit date: | 2019-05-03 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.539 Å) | | Cite: | Lucina pectinata oxyhemoglobin (II-III) heterodimer pH susceptibility.
J.Inorg.Biochem., 207, 2020
|
|
1BKA
 
 | | OXALATE-SUBSTITUTED DIFERRIC LACTOFERRIN | | Descriptor: | FE (III) ION, LACTOFERRIN, OXALATE ION | | Authors: | Baker, H.M, Smith, C.A, Baker, E.N. | | Deposit date: | 1996-04-15 | | Release date: | 1996-11-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Anion binding by transferrins: importance of second-shell effects revealed by the crystal structure of oxalate-substituted diferric lactoferrin.
Biochemistry, 35, 1996
|
|
1JBV
 
 | | FPGS-AMPPCP complex | | Descriptor: | FOLYLPOLYGLUTAMATE SYNTHASE, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Sun, X, Cross, J.A, Bognar, A.L, Baker, E.N, Smith, C.A. | | Deposit date: | 2001-06-06 | | Release date: | 2001-09-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Folate-binding triggers the activation of folylpolyglutamate synthetase.
J.Mol.Biol., 310, 2001
|
|
1JBW
 
 | | FPGS-AMPPCP-folate complex | | Descriptor: | 5,10-METHYLENE-6-HYDROFOLIC ACID, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, FOLYLPOLYGLUTAMATE SYNTHASE, ... | | Authors: | Sun, X, Cross, J.A, Bognar, A.L, Baker, E.N, Smith, C.A. | | Deposit date: | 2001-06-06 | | Release date: | 2001-09-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Folate-binding triggers the activation of folylpolyglutamate synthetase.
J.Mol.Biol., 310, 2001
|
|
1FCK
 
 | | STRUCTURE OF DICERIC HUMAN LACTOFERRIN | | Descriptor: | CARBONATE ION, CERIUM (III) ION, LACTOFERRIN | | Authors: | Baker, H.M, Baker, C.J, Smith, C.A, Baker, E.N. | | Deposit date: | 2000-07-18 | | Release date: | 2001-01-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Metal substitution in transferrins: specific binding of cerium(IV) revealed by the crystal structure of cerium-substituted human lactoferrin.
J.Biol.Inorg.Chem., 5, 2000
|
|
1A8F
 
 | | HUMAN SERUM TRANSFERRIN, RECOMBINANT N-TERMINAL LOBE | | Descriptor: | CARBONATE ION, FE (III) ION, SERUM TRANSFERRIN | | Authors: | Macgillivray, R.T.A, Moore, S.A, Chen, J, Anderson, B.F, Baker, H, Luo, Y, Bewley, M, Smith, C.A, Murphy, M.E.P, Wang, Y, Mason, A.B, Woodworth, R.C, Brayer, G.D, Baker, E.N. | | Deposit date: | 1998-03-25 | | Release date: | 1998-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Two high-resolution crystal structures of the recombinant N-lobe of human transferrin reveal a structural change implicated in iron release.
Biochemistry, 37, 1998
|
|
