4BUZ
 
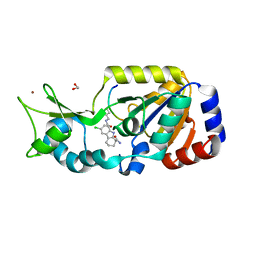 | | SIR2 COMPLEX STRUCTURE MIXTURE OF EX-527 INHIBITOR AND REACTION PRODUCTS OR OF REACTION SUBSTRATES P53 PEPTIDE AND NAD | | Descriptor: | (1S)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1- carboxamide, 1,2-ETHANEDIOL, 2'-O-ACETYL ADENOSINE-5-DIPHOSPHORIBOSE, ... | | Authors: | Weyand, M, Lakshminarasimhan, M, Gertz, M, Steegborn, C. | | Deposit date: | 2013-06-24 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ex-527 Inhibits Sirtuins by Exploiting Their Unique Nad+-Dependent Deacetylation Mechanism
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4IKG
 
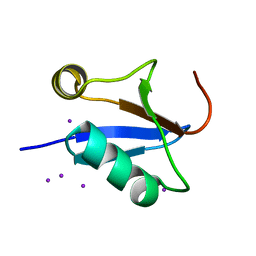 | |
1PD2
 
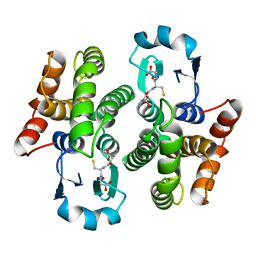 | |
3T9N
 
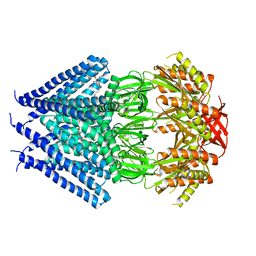 | | Crystal structure of a membrane protein | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Small-conductance mechanosensitive channel | | Authors: | Yang, M, Zhang, X, Ge, J, Wang, J. | | Deposit date: | 2011-08-03 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.456 Å) | | Cite: | Structure and molecular mechanism of an anion-selective mechanosensitive channel of small conductance
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UAL
 
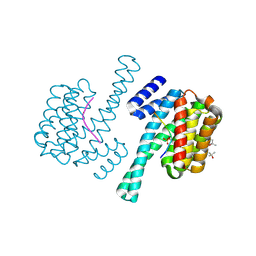 | |
5YE2
 
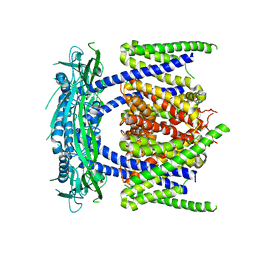 | |
5YE1
 
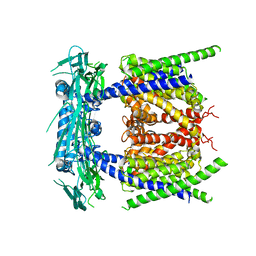 | |
5YE5
 
 | | structure of endo-lysosomal TRPML1 channel inserting into nanodisc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, mammalian endo-lysosomal TRPML1 channel | | Authors: | Yang, M, Gao, N. | | Deposit date: | 2017-09-15 | | Release date: | 2017-11-15 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Cryo-EM structures of the mammalian endo-lysosomal TRPML1 channel elucidate the combined regulation mechanism
Protein Cell, 8, 2017
|
|
5YDZ
 
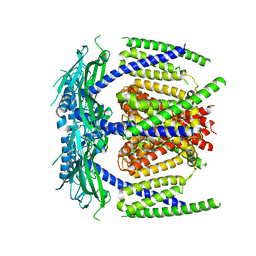 | |
4NQJ
 
 | | Structure of coiled-coil domain | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, E3 ubiquitin-protein ligase TRIM69 | | Authors: | Yang, M, Li, Y. | | Deposit date: | 2013-11-25 | | Release date: | 2014-05-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Structural insights into the TRIM family of ubiquitin E3 ligases.
Cell Res., 24, 2014
|
|
4BGC
 
 | |
2YW9
 
 | |
2ZIY
 
 | | Crystal structure of squid rhodopsin | | Descriptor: | PALMITIC ACID, RETINAL, Rhodopsin | | Authors: | Miyano, M, Shimamura, T. | | Deposit date: | 2008-02-27 | | Release date: | 2008-05-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structure of squid rhodopsin with intracellularly extended cytoplasmic region
J.Biol.Chem., 283, 2008
|
|
7CK6
 
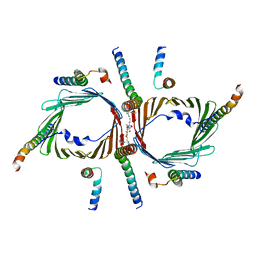 | | Protein translocase of mitochondria | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Mitochondrial import receptor subunit TOM22 homolog, Mitochondrial import receptor subunit TOM40 homolog, ... | | Authors: | Yang, M, Wang, W, Zhang, L, Chen, X. | | Deposit date: | 2020-07-15 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Atomic structure of human TOM core complex.
Cell Discov, 6, 2020
|
|
7E3O
 
 | |
4WA6
 
 | |
4WJA
 
 | | Crystal Structure of PAXX | | Descriptor: | Uncharacterized protein C9orf142 | | Authors: | Xing, M, Yang, M, Huo, W, Feng, F, Wei, L, Ning, S, Yan, Z, Li, W, Wang, Q, Hou, M, Dong, C, Guo, R, Gao, G, Ji, J, Lan, L, Liang, H, Xu, D. | | Deposit date: | 2014-09-29 | | Release date: | 2015-03-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Interactome analysis identifies a new paralogue of XRCC4 in non-homologous end joining DNA repair pathway.
Nat Commun, 6, 2015
|
|
1FPS
 
 | |
1OSX
 
 | | Solution Structure of the Extracellular Domain of BLyS Receptor 3 (BR3) | | Descriptor: | Tumor necrosis factor receptor superfamily member 13C | | Authors: | Gordon, N.C, Pan, B, Hymowitz, S.G, Yin, J.P, Kelley, R.F, Cochran, A.G, Yan, M, Dixit, V.M, Fairbrother, W.J, Starovasnik, M.A. | | Deposit date: | 2003-03-20 | | Release date: | 2003-05-27 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | BAFF/BLyS receptor 3 comprises a minimal TNF receptor-like module that encodes a highly focused ligand-binding site
Biochemistry, 42, 2003
|
|
1OSG
 
 | | Complex between BAFF and a BR3 derived peptide presented in a beta-hairpin scaffold | | Descriptor: | BR3 derived PEPTIDE, MAGNESIUM ION, Tumor necrosis factor ligand superfamily member 13B | | Authors: | Gordon, N.C, Pan, B, Hymowitz, S.G, Yin, J.P, Kelley, R.F, Cochran, A.G, Yan, M, Dixit, V.M, Fairbrother, W.J, Starovasnik, M.A. | | Deposit date: | 2003-03-19 | | Release date: | 2003-05-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | BAFF/BLyS receptor 3 comprises a minimal TNF receptor-like module that encodes a highly focused ligand-binding site
Biochemistry, 42, 2003
|
|
5GPN
 
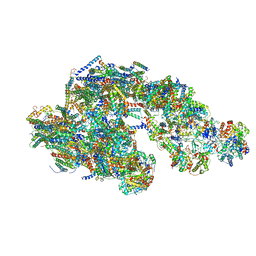 | | Architecture of mammalian respirasome | | Descriptor: | Acyl carrier protein, COPPER (II) ION, Cytochrome b, ... | | Authors: | Gu, J, Wu, M, Guo, R, Yang, M. | | Deposit date: | 2016-08-03 | | Release date: | 2017-04-05 | | Last modified: | 2017-04-12 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | The architecture of the mammalian respirasome.
Nature, 537, 2016
|
|
5ZHZ
 
 | | Crystal structure of the apurinic/apyrimidinic endonuclease IV from Mycobacterium tuberculosis | | Descriptor: | Probable endonuclease 4, SULFATE ION, ZINC ION | | Authors: | Zhang, W, Xu, Y, Yan, M, Li, S, Wang, H, Yang, H, Zhou, W, Rao, Z. | | Deposit date: | 2018-03-13 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Crystal structure of the apurinic/apyrimidinic endonuclease IV from Mycobacterium tuberculosis.
Biochem. Biophys. Res. Commun., 498, 2018
|
|
5ZHY
 
 | | Structural characterization of the HCoV-229E fusion core | | Descriptor: | Spike glycoprotein | | Authors: | Zhang, W, Zheng, Q, Yan, M, Chen, X, Yang, H, Zhou, W, Rao, Z. | | Deposit date: | 2018-03-13 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.441 Å) | | Cite: | Structural characterization of the HCoV-229E fusion core.
Biochem. Biophys. Res. Commun., 497, 2018
|
|
1RJ7
 
 | | Crystal structure of EDA-A1 | | Descriptor: | Ectodysplasin A | | Authors: | Hymowitz, S.G, Compaan, D.M, Yan, M, Ackerly, H, Dixit, V.M, Starovasnik, M.A, de Vos, A.M. | | Deposit date: | 2003-11-18 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structures of EDA-A1 and EDA-A2: splice variants with distinct receptor specificity.
Structure, 11, 2003
|
|
1RJ8
 
 | | The crystal structure of TNF family member EDA-A2 | | Descriptor: | ectodysplasin-A isoform EDA-A2 | | Authors: | Hymowitz, S.G, Compaan, D.M, Yan, M, Ackerly, H, Dixit, V.M, Starovasnik, M.A, de Vos, A.M. | | Deposit date: | 2003-11-18 | | Release date: | 2003-12-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | The Crystal Structure of EDA-A1 and EDA-A2: splice variants with distinct receptor specificity
STRUCTURE, 11, 2003
|
|
