8JYP
 
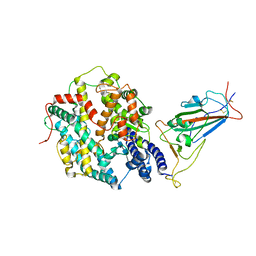 | | Structure of SARS-CoV-2 XBB.1.5 spike RBD in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-07-03 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 XBB.1.5 variant
To Be Published
|
|
8JYN
 
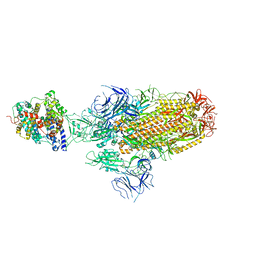 | | Structure of SARS-CoV-2 XBB.1.5 spike glycoprotein in complex with ACE2 (1-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-07-03 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 XBB.1.5 variant
To Be Published
|
|
8JYO
 
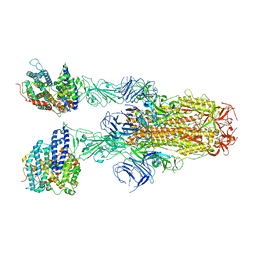 | | Structure of SARS-CoV-2 XBB.1.5 spike glycoprotein in complex with ACE2 (2-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-07-03 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 XBB.1.5 variant
To Be Published
|
|
4IIE
 
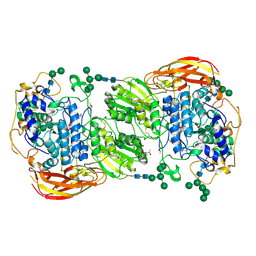 | | Crystal structure of beta-glucosidase 1 from Aspergillus aculeatus in complex with calystegine B(2) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Suzuki, K, Sumitani, J, Kawaguchi, T, Fushinobu, S. | | Deposit date: | 2012-12-20 | | Release date: | 2013-04-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of glycoside hydrolase family 3 beta-glucosidase 1 from Aspergillus aculeatus
Biochem.J., 452, 2013
|
|
6R9W
 
 | | Crystal structure of InhA in complex with AP-124 inhibitor | | Descriptor: | (2~{S})-1-(benzimidazol-1-yl)-3-(2,3-dihydro-1~{H}-inden-5-yloxy)propan-2-ol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Takebayashi, Y, Hinchliffe, P, Spencer, J. | | Deposit date: | 2019-04-04 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of New and Potent InhA Inhibitors as Antituberculosis Agents: Structure-Based Virtual Screening Validated by Biological Assays and X-ray Crystallography.
J.Chem.Inf.Model., 60, 2020
|
|
4IIF
 
 | | Crystal structure of beta-glucosidase 1 from Aspergillus aculeatus in complex with castanospermine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Suzuki, K, Sumitani, J, Kawaguchi, T, Fushinobu, S. | | Deposit date: | 2012-12-20 | | Release date: | 2013-04-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structures of glycoside hydrolase family 3 beta-glucosidase 1 from Aspergillus aculeatus
Biochem.J., 452, 2013
|
|
5L3I
 
 | |
4IIH
 
 | | Crystal structure of beta-glucosidase 1 from Aspergillus aculeatus in complex with thiocellobiose | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Suzuki, K, Sumitani, J, Kawaguchi, T, Fushinobu, S. | | Deposit date: | 2012-12-20 | | Release date: | 2013-04-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of glycoside hydrolase family 3 beta-glucosidase 1 from Aspergillus aculeatus
Biochem.J., 452, 2013
|
|
4IIB
 
 | | Crystal structure of beta-glucosidase 1 from Aspergillus aculeatus | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Suzuki, K, Sumitani, J, Kawaguchi, T, Fushinobu, S. | | Deposit date: | 2012-12-20 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of glycoside hydrolase family 3 beta-glucosidase 1 from Aspergillus aculeatus
Biochem.J., 452, 2013
|
|
4IID
 
 | | Crystal structure of beta-glucosidase 1 from Aspergillus aculeatus in complex with 1-deoxynojirimycin | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 1-DEOXYNOJIRIMYCIN, ... | | Authors: | Suzuki, K, Sumitani, J, Kawaguchi, T, Fushinobu, S. | | Deposit date: | 2012-12-20 | | Release date: | 2013-04-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of glycoside hydrolase family 3 beta-glucosidase 1 from Aspergillus aculeatus
Biochem.J., 452, 2013
|
|
4IIG
 
 | | Crystal structure of beta-glucosidase 1 from Aspergillus aculeatus in complex with D-glucose | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Suzuki, K, Sumitani, J, Kawaguchi, T, Fushinobu, S. | | Deposit date: | 2012-12-20 | | Release date: | 2013-04-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of glycoside hydrolase family 3 beta-glucosidase 1 from Aspergillus aculeatus
Biochem.J., 452, 2013
|
|
4IIC
 
 | | Crystal structure of beta-glucosidase 1 from Aspergillus aculeatus in complex with isofagomine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Suzuki, K, Sumitani, J, Kawaguchi, T, Fushinobu, S. | | Deposit date: | 2012-12-20 | | Release date: | 2013-04-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of glycoside hydrolase family 3 beta-glucosidase 1 from Aspergillus aculeatus
Biochem.J., 452, 2013
|
|
3VNO
 
 | | Cytochrome P450SP alpha (CYP152B1) mutant R241E | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Fatty acid alpha-hydroxylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fujishiro, T, Shoji, O, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2012-01-17 | | Release date: | 2013-02-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | A substrate-binding-state mimic of H2O2-dependent cytochrome P450 produced by one-point mutagenesis and peroxygenation of non-native substrates
Catalysis Science And Technology, 6, 2016
|
|
3VTJ
 
 | | Cytochrome P450SP alpha (CYP152B1) mutant A245H | | Descriptor: | Fatty acid alpha-hydroxylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fujishiro, T, Shoji, O, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2012-05-30 | | Release date: | 2013-06-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | A substrate-binding-state mimic of H2O2-dependent cytochrome P450 produced by one-point mutagenesis and peroxygenation of non-native substrates
Catalysis Science And Technology, 6, 2016
|
|
6N6R
 
 | |
6N6S
 
 | | Crystal structure of ABIN-1 UBAN | | Descriptor: | TNFAIP3-interacting protein 1 | | Authors: | Rahighi, S, Dikic, I, Wakatsuki, S. | | Deposit date: | 2018-11-27 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular Recognition of M1-Linked Ubiquitin Chains by Native and Phosphorylated UBAN Domains.
J.Mol.Biol., 431, 2019
|
|
7PP0
 
 | | Crystal structure of the VIM-2 acquired metallo-beta-Lactamase in complex with compound 28 (JMV-7038) | | Descriptor: | 2-[2-[3-[3-(2-morpholin-4-ylethoxy)phenyl]-5-sulfanylidene-1H-1,2,4-triazol-4-yl]ethyl]benzoic acid, ACETATE ION, Metallo-beta-lactamase VIM-2-like protein, ... | | Authors: | Tassone, G, Benvenuti, M, Verdirosa, F, Corsica, G, Chelini, G, De Luca, F, Docquier, J.D, Pozzi, C, Mangani, S. | | Deposit date: | 2021-09-10 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | 1,2,4-Triazole-3-Thione Analogues with a 2-Ethylbenzoic Acid at Position 4 as VIM-type Metallo-beta-Lactamase Inhibitors.
Chemmedchem, 17, 2022
|
|
6N5M
 
 | |
5XF3
 
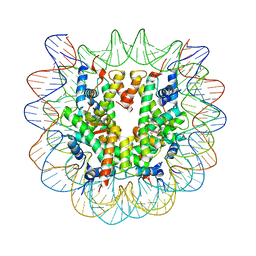 | | Nucleosome core particle with an adduct of a binuclear RAPTA (Ru-arene-phosphaadamantane) compound having a 1,2-diphenylethylenediamine linker (R,R-configuration) | | Descriptor: | (1R,2R)-1,2-diphenylethane-1,2-diamine, DNA (145-MER), Histone H2A type 1-B/E, ... | | Authors: | Ma, Z, Adhireksan, Z, Murray, B.S, Dyson, P.J, Davey, C.A. | | Deposit date: | 2017-04-07 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Nucleosome acidic patch-targeting binuclear ruthenium compounds induce aberrant chromatin condensation
Nat Commun, 8, 2017
|
|
5XDP
 
 | |
5XF5
 
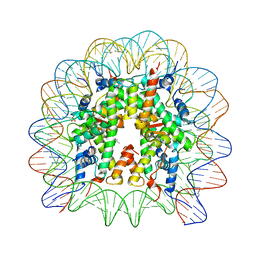 | | Nucleosome core particle with an adduct of a binuclear RAPTA (Ru-arene-phosphaadamantane) compound having a 1,2-diphenylethylenediamine linker (R,S-configuration) | | Descriptor: | (1S,2R)-1,2-diphenylethane-1,2-diamine, DNA (145-MER), Histone H2A type 1-B/E, ... | | Authors: | Ma, Z, Adhireksan, Z, Murray, B.S, Dyson, P.J, Davey, C.A. | | Deposit date: | 2017-04-07 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Nucleosome acidic patch-targeting binuclear ruthenium compounds induce aberrant chromatin condensation
Nat Commun, 8, 2017
|
|
8GBU
 
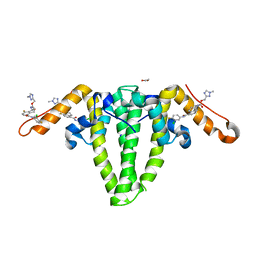 | | Hepatitis B capsid Y132A mutant with compound AB-506 | | Descriptor: | (1-methyl-1H-1,2,4-triazol-3-yl)methyl {(1S)-4-[(3-chloro-4-fluorophenyl)carbamoyl]-7-fluoro-2,3-dihydro-1H-inden-1-yl}carbamate, 1,2-ETHANEDIOL, Capsid protein | | Authors: | Horanyi, P.S, Mayclin, S.J. | | Deposit date: | 2023-02-28 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design, synthesis, and structure-activity relationship of a bicyclic HBV capsid assembly modulator chemotype leading to the identification of clinical candidate AB-506.
Bioorg.Med.Chem.Lett., 94, 2023
|
|
5XF6
 
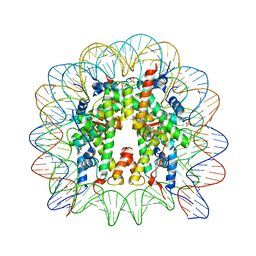 | | Nucleosome core particle with an adduct of a binuclear RAPTA (Ru-arene-phosphaadamantane) compound having an ethylenediamine linker | | Descriptor: | DNA (145-MER), ETHANE-1,2-DIAMINE, Histone H2A, ... | | Authors: | Ma, Z, Adhireksan, Z, Murray, B.S, Dyson, P.J, Davey, C.A. | | Deposit date: | 2017-04-07 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Nucleosome acidic patch-targeting binuclear ruthenium compounds induce aberrant chromatin condensation
Nat Commun, 8, 2017
|
|
5XF4
 
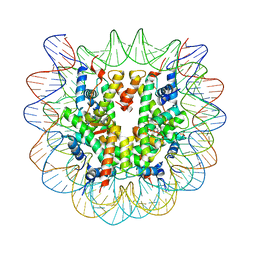 | | Nucleosome core particle with an adduct of a binuclear RAPTA (Ru-arene-phosphaadamantane) compound having a 1,2-diphenylethylenediamine linker (S,S-configuration) | | Descriptor: | (1S,2S)-1,2-diphenylethane-1,2-diamine, DNA (145-MER), Histone H2A type 1-B/E, ... | | Authors: | Ma, Z, Adhireksan, Z, Murray, B.S, Dyson, P.J, Davey, C.A. | | Deposit date: | 2017-04-07 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Nucleosome acidic patch-targeting binuclear ruthenium compounds induce aberrant chromatin condensation
Nat Commun, 8, 2017
|
|
8JU3
 
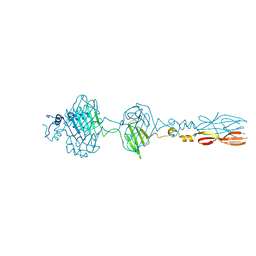 | | Mu phage tail fiber | | Descriptor: | Tail fiber protein S,Tail fiber protein S' | | Authors: | Yamashita, E, Takeda, S. | | Deposit date: | 2023-06-24 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Determination of the three-dimensional structure of bacteriophage Mu(-) tail fiber and its characterization.
Virology, 593, 2024
|
|
