6PBH
 
 | |
4G57
 
 | | Staphylococcal Nuclease double mutant I72L, I92L | | Descriptor: | Thermonuclease | | Authors: | Sakon, J, Stites, W.E, Gill, E, Latimer, E, Rosser, J. | | Deposit date: | 2012-07-17 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: |
|
|
6SV1
 
 | |
6SUW
 
 | |
7SNE
 
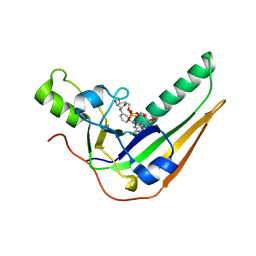 | | Pertussis toxin S1 subunit bound to BaAD | | Descriptor: | Pertussis toxin subunit 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl [(2R,3S,4R,5S)-5-(3-carbamoylanilino)-3,4-dihydroxyoxolan-2-yl]methyl dihydrogen diphosphate (non-preferred name) | | Authors: | Littler, D.R, Beddoe, T. | | Deposit date: | 2021-10-28 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.00011 Å) | | Cite: | Crystal structures of pertussis toxin with NAD + and analogs provide structural insights into the mechanism of its cytosolic ADP-ribosylation activity.
J.Biol.Chem., 298, 2022
|
|
7AV7
 
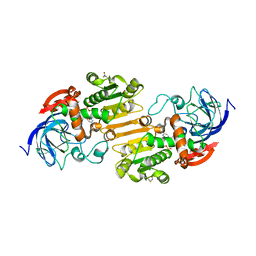 | | Crystal structure of S-nitrosylated nitrosoglutathione reductase(GSNOR)from Chlamydomonas reinhardtii, in complex with NAD+ | | Descriptor: | CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, S-(hydroxymethyl)glutathione dehydrogenase, ... | | Authors: | Fermani, S, Zaffagnini, M, Falini, G, Lemaire, S.D. | | Deposit date: | 2020-11-04 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional insights into nitrosoglutathione reductase from Chlamydomonas reinhardtii.
Redox Biol, 38, 2020
|
|
8ESD
 
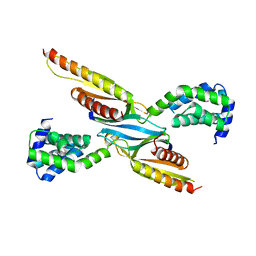 | | Crystal structure of COMMD7-COMMD9-COMMD5-COMMD10 tetramer | | Descriptor: | COMM domain-containing protein 10, COMM domain-containing protein 5, COMM domain-containing protein 7, ... | | Authors: | Healy, M.D, Collins, B.M. | | Deposit date: | 2022-10-13 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Structure of the endosomal Commander complex linked to Ritscher-Schinzel syndrome.
Cell, 186, 2023
|
|
8ESE
 
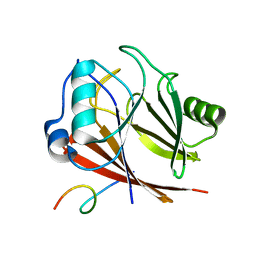 | |
2BK1
 
 | | The pore structure of pneumolysin, obtained by fitting the alpha carbon trace of perfringolysin O into a cryo-EM map | | Descriptor: | PERFRINGOLYSIN O | | Authors: | Tilley, S.J, Orlova, E.V, Gilbert, R.J.C, Andrew, P.W, Saibil, H.R. | | Deposit date: | 2005-02-10 | | Release date: | 2005-05-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (29 Å) | | Cite: | Structural Basis of Pore Formation by the Bacterial Toxin Pneumolysin
Cell(Cambridge,Mass.), 121, 2005
|
|
2BK2
 
 | | The prepore structure of pneumolysin, obtained by fitting the alpha carbon trace of perfringolysin O into a cryo-EM map | | Descriptor: | PERFRINGOLYSIN O | | Authors: | Tilley, S.J, Orlova, E.V, Gilbert, R.J.C, Andrew, P.W, Saibil, H.R. | | Deposit date: | 2005-02-10 | | Release date: | 2005-05-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (28 Å) | | Cite: | Structural Basis of Pore Formation by the Bacterial Toxin Pneumolysin
Cell(Cambridge,Mass.), 121, 2005
|
|
4P15
 
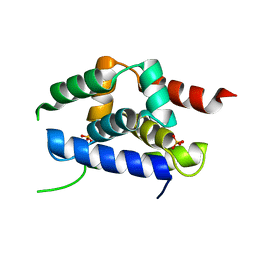 | | Structure of the ClpC N-terminal domain from an alkaliphilic Bacillus lehensis G1 species | | Descriptor: | Bacillus lehensis ClpC N-terminal domain, SULFATE ION | | Authors: | Rashid, S.A, Littler, D.R, Illias, R.M, Murad, A.M.A, Rossjohn, J, Beddoe, T, Mahadi, N.M. | | Deposit date: | 2014-01-31 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the ClpC N-terminal domain at 1.85 Angstroms resolution from an alkaliphilic Bacillus lehensis G1 species
To Be Published
|
|
6NF7
 
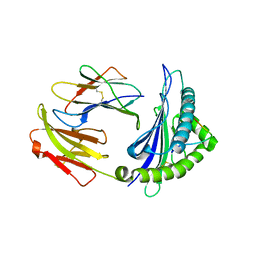 | | Crystal Structure of RT1.Aa-Bu31-10 | | Descriptor: | Beta-2-microglobulin, Bu31-10 peptide, RT1A.a | | Authors: | Gras, S. | | Deposit date: | 2018-12-19 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cross-Reactive Donor-Specific CD8+Tregs Efficiently Prevent Transplant Rejection.
Cell Rep, 29, 2019
|
|
9GSS
 
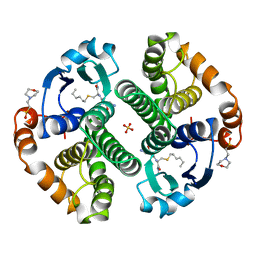 | | HUMAN GLUTATHIONE S-TRANSFERASE P1-1, COMPLEX WITH S-HEXYL GLUTATHIONE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE S-TRANSFERASE P1-1, S-HEXYLGLUTATHIONE, ... | | Authors: | Oakley, A, Parker, M. | | Deposit date: | 1997-08-14 | | Release date: | 1998-09-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The structures of human glutathione transferase P1-1 in complex with glutathione and various inhibitors at high resolution.
J.Mol.Biol., 274, 1997
|
|
6MT4
 
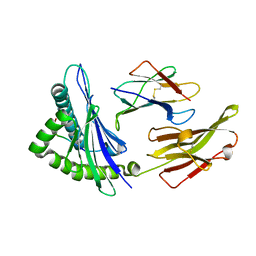 | |
6MTM
 
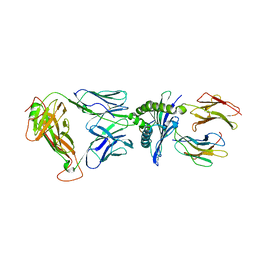 | | Crystal Structure of EM2 TCR in complex with HLA-B*37:01-NP338 | | Descriptor: | Beta-2-microglobulin, EM2 TCR alpha chain, EM2 TCR beta chain, ... | | Authors: | Gras, S. | | Deposit date: | 2018-10-19 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Broad CD8+T cell cross-recognition of distinct influenza A strains in humans.
Nat Commun, 9, 2018
|
|
5KBF
 
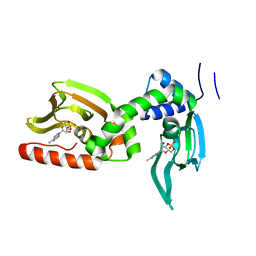 | | cAMP bound PfPKA-R (141-441) | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CAMP-dependent protein kinase regulatory subunit, putative | | Authors: | Littler, D.R, Gilson, P.R, Crabb, B.S, Rossjohn, J.J. | | Deposit date: | 2016-06-03 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Disrupting the Allosteric Interaction between the Plasmodium falciparum cAMP-dependent Kinase and Its Regulatory Subunit.
J. Biol. Chem., 291, 2016
|
|
5K8S
 
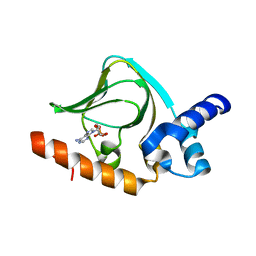 | | cAMP bound PfPKA-R (297-441) | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CAMP-dependent protein kinase regulatory subunit | | Authors: | Littler, D.R, Gilson, P.R, Crabb, B.S, Rossjohn, J.J. | | Deposit date: | 2016-05-31 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Disrupting the Allosteric Interaction between the Plasmodium falciparum cAMP-dependent Kinase and Its Regulatory Subunit.
J. Biol. Chem., 291, 2016
|
|
5D7R
 
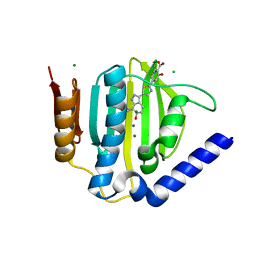 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-hydroxy-5-[5-(6-hydroxy-7-propyl-2H-indazol-3-yl)-1,3-thiazol-2-yl]pyridine-2-carboxylic acid, CHLORIDE ION, ... | | Authors: | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | Deposit date: | 2015-08-14 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of Indazole Derivatives as a Novel Class of Bacterial Gyrase B Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
5D7D
 
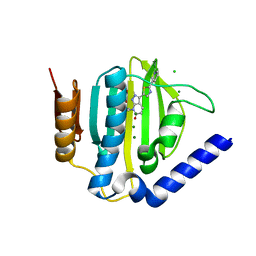 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 7-propyl-3-[2-(pyridin-3-yl)-1,3-thiazol-5-yl]-1,7-dihydro-6H-pyrazolo[3,4-b]pyridin-6-one, CHLORIDE ION, ... | | Authors: | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | Deposit date: | 2015-08-13 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Indazole Derivatives as a Novel Class of Bacterial Gyrase B Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
3OF6
 
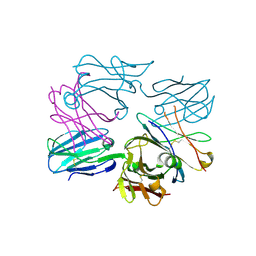 | | Human pre-T cell receptor crystal structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Pre T-cell antigen receptor alpha, T cell receptor beta chain | | Authors: | Pang, S.S. | | Deposit date: | 2010-08-13 | | Release date: | 2010-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural basis for autonomous dimerization of the pre-T-cell antigen receptor
Nature, 467, 2010
|
|
9ENJ
 
 | | L-amino acid oxidase 4 (HcLAAO4) from the fungus Hebeloma cylindrosporum in complex with L-glutamate | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, GLUTAMIC ACID, L-amino acid oxidase 4, ... | | Authors: | Gilzer, D, Koopmeiners, S, Fischer von Mollard, G, Niemann, H.H. | | Deposit date: | 2024-03-13 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and enzyme engineering of the broad substrate spectrum l-amino acid oxidase 4 from the fungus Hebeloma cylindrosporum.
Febs Lett., 2024
|
|
9ENK
 
 | | L-amino acid oxidase 4 (HcLAAO4) from the fungus Hebeloma cylindrosporum in complex with L-phenylalanine | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, L-amino acid oxidase 4, PHENYLALANINE, ... | | Authors: | Gilzer, D, Koopmeiners, S, Fischer von Mollard, G, Niemann, H.H. | | Deposit date: | 2024-03-13 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and enzyme engineering of the broad substrate spectrum l-amino acid oxidase 4 from the fungus Hebeloma cylindrosporum.
Febs Lett., 2024
|
|
9ENI
 
 | | L-amino acid oxidase 4 (HcLAAO4) from the fungus Hebeloma cylindrosporum in complex with L-glutamine | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, GLUTAMINE, L-amino acid oxidase 4, ... | | Authors: | Gilzer, D, Koopmeiners, S, Fischer von Mollard, G, Niemann, H.H. | | Deposit date: | 2024-03-13 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and enzyme engineering of the broad substrate spectrum l-amino acid oxidase 4 from the fungus Hebeloma cylindrosporum.
Febs Lett., 2024
|
|
9ENH
 
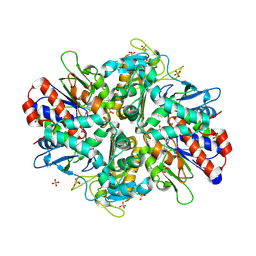 | | L-amino acid oxidase 4 (HcLAAO4) from the fungus Hebeloma cylindrosporum | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-amino acid oxidase 4, S-1,2-PROPANEDIOL, ... | | Authors: | Gilzer, D, Koopmeiners, S, Fischer von Mollard, G, Niemann, H.H. | | Deposit date: | 2024-03-13 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and enzyme engineering of the broad substrate spectrum l-amino acid oxidase 4 from the fungus Hebeloma cylindrosporum.
Febs Lett., 2024
|
|
9ENN
 
 | | L-amino acid oxidase 4 (HcLAAO4) from the fungus Hebeloma cylindrosporum in complex with N-epsilon-acetyl-L-lysine | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, L-amino acid oxidase 4, N(6)-ACETYLLYSINE, ... | | Authors: | Gilzer, D, Koopmeiners, S, Fischer von Mollard, G, Niemann, H.H. | | Deposit date: | 2024-03-13 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and enzyme engineering of the broad substrate spectrum l-amino acid oxidase 4 from the fungus Hebeloma cylindrosporum.
Febs Lett., 2024
|
|
