6WBD
 
 | |
6XNQ
 
 | |
3NRR
 
 | |
3OC9
 
 | |
3P32
 
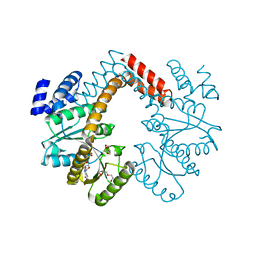 | |
5B8I
 
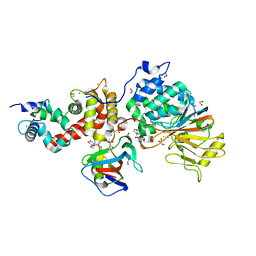 | | Crystal structure of Calcineurin A and Calcineurin B in complex with FKBP12 and FK506 from Coccidioides immitis RS | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID), Fox III, D, Dranow, D.M, Lorimer, D.D, Edwards, T.E. | | Deposit date: | 2015-05-03 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Harnessing calcineurin-FK506-FKBP12 crystal structures from invasive fungal pathogens to develop antifungal agents.
Nat Commun, 10, 2019
|
|
7U0S
 
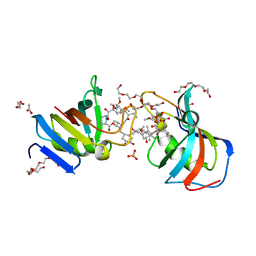 | | Crystal Structure of FK506-binding protein 1A from Aspergillus fumigatus Bound to Ascomycin | | Descriptor: | (3S,4R,5S,8R,9E,12S,14S,15R,16S,18R,19R,22R,26aS)-8-ethyl-5,19-dihydroxy-3-{(1E)-1-[(1R,3R,4R)-4-hydroxy-3-methoxycyclohexyl]prop-1-en-2-yl}-14,16-dimethoxy-4,10,12,18-tetramethyl-5,6,8,11,12,13,14,15,16,17,18,19,24,25,26,26a-hexadecahydro-3H-15,19-epoxypyrido[2,1-c][1,4]oxazacyclotricosine-1,7,20,21(4H,23H)-tetrone, ACETATE ION, FK506-binding protein 1A, ... | | Authors: | DeBouver, N.D, Fox III, D, Hoy, M.J, Heitman, J, Lorimer, D.D, Horanyi, P.S, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-02-18 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Guided Synthesis of FK506 and FK520 Analogs with Increased Selectivity Exhibit In Vivo Therapeutic Efficacy against Cryptococcus.
Mbio, 13, 2022
|
|
7U0U
 
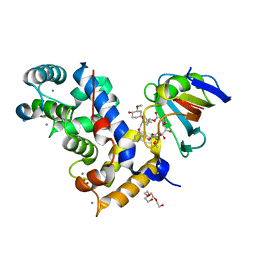 | | Crystal Structure of a Aspergillus fumigatus Calcineurin A - Calcineurin B fusion bound to FKBP12 and FK-506 | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Fox III, D, Abendroth, J, DeBouver, N.D, Hoy, M.J, Heitman, J, Lorimer, D.D, Horanyi, P.S, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-02-18 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Synthesis of FK506 and FK520 Analogs with Increased Selectivity Exhibit In Vivo Therapeutic Efficacy against Cryptococcus.
Mbio, 13, 2022
|
|
7U0T
 
 | | Crystal Structure of a human Calcineurin A - Calcineurin B fusion bound to FKBP12 and FK-520 | | Descriptor: | (3S,4R,5S,8R,9E,12S,14S,15R,16S,18R,19R,22R,26aS)-8-ethyl-5,19-dihydroxy-3-{(1E)-1-[(1R,3R,4R)-4-hydroxy-3-methoxycyclohexyl]prop-1-en-2-yl}-14,16-dimethoxy-4,10,12,18-tetramethyl-5,6,8,11,12,13,14,15,16,17,18,19,24,25,26,26a-hexadecahydro-3H-15,19-epoxypyrido[2,1-c][1,4]oxazacyclotricosine-1,7,20,21(4H,23H)-tetrone, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Fox III, D, Mayclin, S.J, DeBouver, N.D, Hoy, M.J, Heitman, J, Lorimer, D.D, Horanyi, P.S, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-02-18 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-Guided Synthesis of FK506 and FK520 Analogs with Increased Selectivity Exhibit In Vivo Therapeutic Efficacy against Cryptococcus.
Mbio, 13, 2022
|
|
6MOM
 
 | | Crystal structure of human Interleukin-1 receptor associated Kinase 4 (IRAK 4, CID 100300) in complex with compound NCC00371481 (BSI 107591) | | Descriptor: | 1,2-ETHANEDIOL, 6-[7-methoxy-6-(1-methyl-1H-pyrazol-4-yl)imidazo[1,2-a]pyridin-3-yl]-N-[(3R)-pyrrolidin-3-yl]pyridin-2-amine, Interleukin-1 receptor-associated kinase 4 | | Authors: | Abendroth, J, Mayclin, S.J, Lorimer, D.D, Starczynowski, D, Hoyt, S, Tawa, G, Thomas, C. | | Deposit date: | 2018-10-04 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Overcoming adaptive therapy resistance in AML by targeting immune response pathways.
Sci Transl Med, 11, 2019
|
|
6MAI
 
 | |
6MFK
 
 | |
6MAO
 
 | |
6MG6
 
 | |
6MDY
 
 | |
6MFM
 
 | |
6MC0
 
 | |
6MFU
 
 | |
6MKE
 
 | |
6MJN
 
 | |
6MJ9
 
 | |
6MWJ
 
 | |
6ML8
 
 | |
6MWK
 
 | |
6MYA
 
 | |
