6J8Y
 
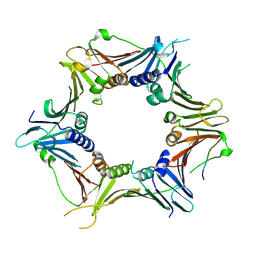 | | Crystal structure of the human RAD9-HUS1-RAD1-RHINO complex | | Descriptor: | Cell cycle checkpoint control protein RAD9A, Cell cycle checkpoint protein RAD1, Checkpoint protein HUS1, ... | | Authors: | Hara, K, Iida, N, Sakurai, H, Hashimoto, H. | | Deposit date: | 2019-01-21 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the RAD9-RAD1-HUS1 checkpoint clamp bound to RHINO sheds light on the other side of the DNA clamp.
J.Biol.Chem., 295, 2020
|
|
7WIS
 
 | |
7WIR
 
 | |
7WKI
 
 | |
3W80
 
 | |
3W7Y
 
 | |
7WYR
 
 | | Crystal structure of Cypovirus Polyhedra mutant fused with CLN025 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Polyhedrin fused with CLN025 | | Authors: | Kojima, M, Abe, S, Hirata, K, Yamashita, K, Ueno, T. | | Deposit date: | 2022-02-16 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Engineering of an in-cell protein crystal for fastening a metastable conformation of a target miniprotein.
Biomater Sci, 11, 2023
|
|
7XEJ
 
 | | SufS with D-cysteine for 5 min | | Descriptor: | (2~{S})-2-[(~{E})-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]-3-sulfanyl-propanoic acid, 1,2-ETHANEDIOL, Cysteine desulfurase SufS, ... | | Authors: | Nakamiura, R, Fujishiro, T. | | Deposit date: | 2022-03-31 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | X-ray crystallographic snapshots of the thioazolidine formation upon the PLP during inhibition of SufS by D-cysteine
to be published
|
|
7XEK
 
 | | SufS with D-cysteine for 6 h | | Descriptor: | (2~{S},4~{S})-2-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]-1,3-thiazolidine-4-carboxylic acid, 1,2-ETHANEDIOL, Cysteine desulfurase SufS, ... | | Authors: | Nakamura, R, Fujishiro, T. | | Deposit date: | 2022-03-31 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | X-ray crystallographic snapshots of the thioazolidine formation upon the PLP during inhibition of SufS by D-cysteine
to be published
|
|
7XEN
 
 | | SufS with L-penicillamine | | Descriptor: | (2~{R})-3-methyl-2-[(~{E})-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]-3-sulfanyl-butanoic acid, Cysteine desulfurase SufS | | Authors: | Nakamura, R, Fujishiro, T. | | Deposit date: | 2022-03-31 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | X-ray crystallographic snapshots of the thioazolidine formation upon the PLP during inhibition of SufS by D-cysteine
to be published
|
|
7XET
 
 | | SufS with beta-cyano-L-alanine | | Descriptor: | (2~{E})-3-cyano-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylimino]propanoic acid, Cysteine desulfurase SufS, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nakamura, R, Fujishiro, T. | | Deposit date: | 2022-03-31 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Inhibition mechanism of SufS by L-propargylglycine
to be published
|
|
7XEP
 
 | | SufS with L-propargylglycine | | Descriptor: | 1,2-ETHANEDIOL, Cysteine desulfurase SufS, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nakamura, R, Fujishiro, T. | | Deposit date: | 2022-03-31 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Inhibition mechanism of SufS by L-propargylglycine
to be published
|
|
7XES
 
 | | NifS with L-penicillamine | | Descriptor: | (2~{R})-3-methyl-2-[(~{E})-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]-3-sulfanyl-butanoic acid, CHLORIDE ION, Cysteine desulfurase IscS, ... | | Authors: | Nakamura, R, Fujishiro, T. | | Deposit date: | 2022-03-31 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray crystallographic snapshots of the thioazolidine formation upon the PLP during inhibition of SufS by D-cysteine
to be published
|
|
7XEQ
 
 | | NifS with D-cysteine | | Descriptor: | (2~{S},4~{S})-2-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]-1,3-thiazolidine-4-carboxylic acid, CHLORIDE ION, Cysteine desulfurase IscS, ... | | Authors: | Nakamura, R, Fujishiro, T. | | Deposit date: | 2022-03-31 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray crystallographic snapshots of the thioazolidine formation upon the PLP during inhibition of SufS by D-cysteine
to be published
|
|
7XEL
 
 | | SufS soaked with D-penicillamine | | Descriptor: | 1,2-ETHANEDIOL, Cysteine desulfurase SufS, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nakamura, R, Fujishiro, T. | | Deposit date: | 2022-03-31 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystallographic snapshots of the thioazolidine formation upon the PLP during inhibition of SufS by D-cysteine
to be published
|
|
6AHP
 
 | | Structure of the 58-167 fragment of FliL | | Descriptor: | Flagellar protein FliL | | Authors: | Takekawa, N, Isumi, M, Sakuma, M, Kojima, S, Homma, M, Imada, K. | | Deposit date: | 2018-08-20 | | Release date: | 2019-03-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure ofVibrioFliL, a New Stomatin-like Protein That Assists the Bacterial Flagellar Motor Function.
MBio, 10, 2019
|
|
6AHQ
 
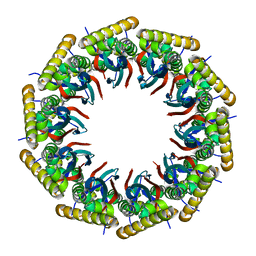 | | Structure of the 40-167 fragment of FliL | | Descriptor: | Flagellar protein FliL | | Authors: | Takekawa, N, Isumi, M, Sakuma, M, Kojima, S, Homma, M, Imada, K. | | Deposit date: | 2018-08-20 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.398 Å) | | Cite: | Structure ofVibrioFliL, a New Stomatin-like Protein That Assists the Bacterial Flagellar Motor Function.
MBio, 10, 2019
|
|
3WMT
 
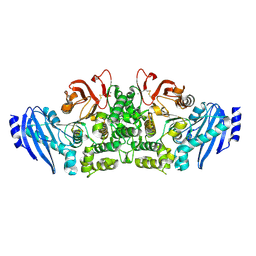 | | Crystal structure of feruloyl esterase B from Aspergillus oryzae | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Probable feruloyl esterase B-1 | | Authors: | Suzuki, K, Ishida, T, Igarashi, K, Koseki, T, Fushinobu, S. | | Deposit date: | 2013-11-25 | | Release date: | 2014-08-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a feruloyl esterase belonging to the tannase family: a disulfide bond near a catalytic triad.
Proteins, 82, 2014
|
|
7YB3
 
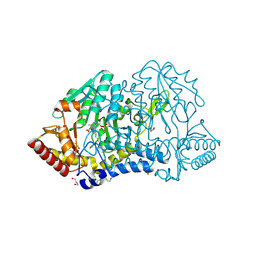 | | SufS with D-cysteine for 1 min | | Descriptor: | Cysteine desulfurase SufS, DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL | | Authors: | Nakamura, R, Fujishiro, T. | | Deposit date: | 2022-06-28 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Snapshots of thiazolidine fomration of PLP-dependent enzyme SufS
to be published
|
|
5VEB
 
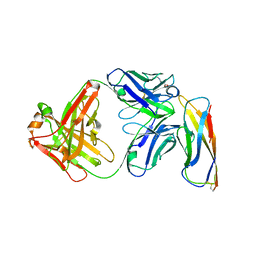 | | Crystal structure of a Fab binding to extracellular domain 5 of Cadherin-6 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, Cadherin-6, anti-CDH6 Fab heavy chain, ... | | Authors: | Zhu, X, Bialucha, C.U, London, A, Clark, K, Hu, T. | | Deposit date: | 2017-04-04 | | Release date: | 2017-06-07 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Discovery and Optimization of HKT288, a Cadherin-6-Targeting ADC for the Treatment of Ovarian and Renal Cancers.
Cancer Discov, 7, 2017
|
|
1A6Y
 
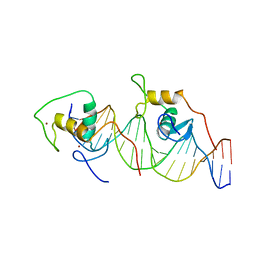 | | REVERBA ORPHAN NUCLEAR RECEPTOR/DNA COMPLEX | | Descriptor: | DNA (5'-D(*CP*AP*AP*CP*TP*AP*GP*GP*TP*CP*AP*CP*(5IT)P*AP*GP*GP*TP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*AP*CP*CP*TP*AP*GP*TP*GP*AP*CP*CP*TP*AP*GP*TP*TP*G)-3'), ORPHAN NUCLEAR RECEPTOR NR1D1, ... | | Authors: | Zhao, Q, Khorasanizadeh, S, Rastinejad, F. | | Deposit date: | 1998-03-04 | | Release date: | 1998-10-21 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural elements of an orphan nuclear receptor-DNA complex.
Mol.Cell, 1, 1998
|
|
5ZO9
 
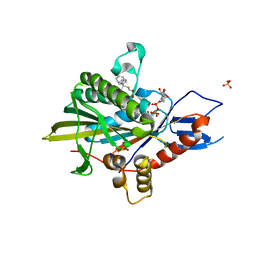 | | Eg5 motor domain in complex with STLC-type inhibitor PVEI0021 (C2 type) | | Descriptor: | (2R)-2-azanyl-3-[(4-methoxyphenyl)-diphenyl-methyl]sulfanyl-propanoic acid, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Yokoyama, H, Sato, K. | | Deposit date: | 2018-04-12 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Thermodynamic Basis of the Enhanced Interaction between Kinesin Spindle Protein Eg5 and STLC-type Inhibitors.
Acs Omega, 3, 2018
|
|
5ZO8
 
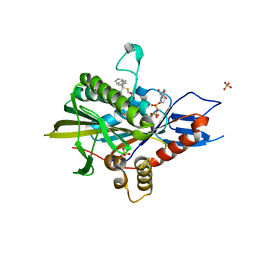 | | Eg5 motor domain in complex with STLC-type inhibitor PVEI0021 (P21 type) | | Descriptor: | (2R)-2-azanyl-3-[(4-methoxyphenyl)-diphenyl-methyl]sulfanyl-propanoic acid, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Yokoyama, H, Sato, K. | | Deposit date: | 2018-04-12 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Thermodynamic Basis of the Enhanced Interaction between Kinesin Spindle Protein Eg5 and STLC-type Inhibitors.
Acs Omega, 3, 2018
|
|
5ZFP
 
 | |
5ZO7
 
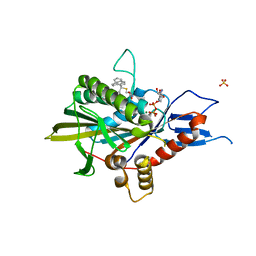 | | Kinesin spindle protein Eg5 in complex with STLC-type inhibitor PVEI0138 | | Descriptor: | (2R)-2-azanyl-3-[[2-(4-methoxyphenyl)-2-tricyclo[9.4.0.0^{3,8}]pentadeca-1(11),3,5,7,12,14-hexaenyl]sulfanyl]propanoic acid, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Yokoyama, H, Sato, K. | | Deposit date: | 2018-04-12 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Thermodynamic Basis of the Enhanced Interaction between Kinesin Spindle Protein Eg5 and STLC-type Inhibitors.
Acs Omega, 3, 2018
|
|
