2RPQ
 
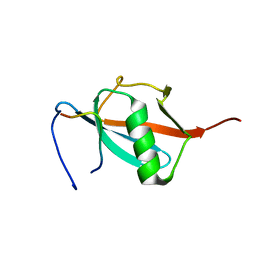 | | Solution Structure of a SUMO-interacting motif of MBD1-containing chromatin-associated factor 1 bound to SUMO-3 | | Descriptor: | Activating transcription factor 7-interacting protein 1, Small ubiquitin-related modifier 2 | | Authors: | Sekiyama, N, Ikegami, T, Yamane, T, Ikeguchi, M, Uchimura, Y, Baba, D, Ariyoshi, M, Tochio, H, Saitoh, H, Shirakawa, M. | | Deposit date: | 2008-07-07 | | Release date: | 2008-10-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the small ubiquitin-like modifier (SUMO)-interacting motif of MBD1-containing chromatin-associated factor 1 bound to SUMO-3
J.Biol.Chem., 283, 2008
|
|
5ZIB
 
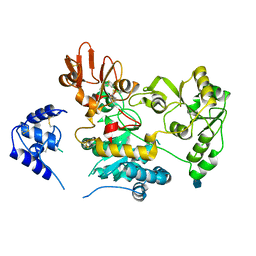 | | Crystal structure of human GnT-V luminal domain in apo form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase A | | Authors: | Nagae, M, Yamaguchi, Y. | | Deposit date: | 2018-03-14 | | Release date: | 2018-08-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of cancer-associated N-acetylglucosaminyltransferase-V.
Nat Commun, 9, 2018
|
|
1GIR
 
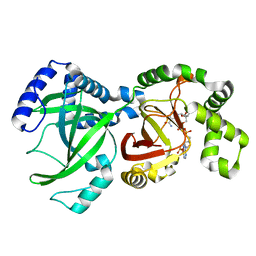 | | CRYSTAL STRUCTURE OF THE ENZYMATIC COMPONET OF IOTA-TOXIN FROM CLOSTRIDIUM PERFRINGENS WITH NADPH | | Descriptor: | IOTA TOXIN COMPONENT IA, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Tsuge, H, Nagahama, M, Nishimura, H, Hisatsune, J, Sakaguchi, Y, Itogawa, Y, Katunuma, N, Sakurai, J. | | Deposit date: | 2001-03-12 | | Release date: | 2003-01-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Site-directed Mutagenesis of Enzymatic Components from Clostridium perfringens Iota-toxin
J.MOL.BIOL., 325, 2003
|
|
1GIQ
 
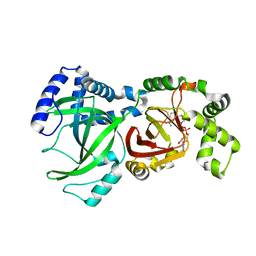 | | Crystal Structure of the Enzymatic Componet of Iota-Toxin from Clostridium Perfringens with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, IOTA TOXIN COMPONENT IA | | Authors: | Tsuge, H, Nagahama, M, Nishimura, H, Hisatsune, J, Sakaguchi, Y, Itogawa, Y, Katunuma, N, Sakurai, J. | | Deposit date: | 2001-03-12 | | Release date: | 2003-01-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure and Site-directed Mutagenesis of Enzymatic Components from Clostridium perfringens Iota-toxin
J.MOL.BIOL., 325, 2003
|
|
5ZIC
 
 | | Crystal structure of human GnT-V luminal domain in complex with acceptor sugar | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-6-thio-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose, Alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase A | | Authors: | Nagae, M, Yamaguchi, Y. | | Deposit date: | 2018-03-14 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and mechanism of cancer-associated N-acetylglucosaminyltransferase-V.
Nat Commun, 9, 2018
|
|
4YSN
 
 | | Structure of aminoacid racemase in complex with PLP | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Putative 4-aminobutyrate aminotransferase | | Authors: | Sakuraba, H, Mutaguchi, Y, Hayashi, J, Ohshima, T. | | Deposit date: | 2015-03-17 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of the novel amino-acid racemase isoleucine 2-epimerase from Lactobacillus buchneri.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5B0O
 
 | | Structure of the FliH-FliI complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Flagellar assembly protein FliH, Flagellum-specific ATP synthase | | Authors: | Imada, K, Uchida, Y, Kinoshita, M, Namba, K, Minamino, T. | | Deposit date: | 2015-11-02 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Insight into the flagella type III export revealed by the complex structure of the type III ATPase and its regulator
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3AQZ
 
 | | Crystal structure of Plodia interpunctella beta-GRP/GNBP3 N-terminal domain with laminarihexaoses | | Descriptor: | Beta-1,3-glucan-binding protein, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Kanagawa, M, Satoh, T, Ikeda, A, Adachi, Y, Ohno, N, Yamaguchi, Y. | | Deposit date: | 2010-11-22 | | Release date: | 2011-06-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into recognition of triple-helical beta-glucans by an insect fungal receptor
J.Biol.Chem., 286, 2011
|
|
3AQY
 
 | | Crystal structure of Plodia interpunctella beta-GRP/GNBP3 N-terminal domain | | Descriptor: | Beta-1,3-glucan-binding protein | | Authors: | Kanagawa, M, Satoh, T, Ikeda, A, Adachi, Y, Ohno, N, Yamaguchi, Y. | | Deposit date: | 2010-11-22 | | Release date: | 2011-06-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural insights into recognition of triple-helical beta-glucans by an insect fungal receptor
J.Biol.Chem., 286, 2011
|
|
5XF9
 
 | | Crystal structure of NAD+-reducing [NiFe]-hydrogenase in the air-oxidized state | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Shomura, Y, Taketa, M, Nakashima, H, Tai, H, Nakagawa, H, Ikeda, Y, Ishii, M, Igarashi, Y, Nishihara, H, Yoon, K.S, Ogo, S, Hirota, S, Higuchi, Y. | | Deposit date: | 2017-04-09 | | Release date: | 2017-08-23 | | Last modified: | 2017-09-20 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural basis of the redox switches in the NAD(+)-reducing soluble [NiFe]-hydrogenase
Science, 357, 2017
|
|
3KLJ
 
 | | Crystal structure of NADH:rubredoxin oxidoreductase from Clostridium acetobutylicum | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NAD(FAD)-dependent dehydrogenase, NirB-family (N-terminal domain) | | Authors: | Nishikawa, K, Shomura, Y, Kawasaki, S, Niimura, Y, Higuchi, Y. | | Deposit date: | 2009-11-08 | | Release date: | 2010-02-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of NADH:rubredoxin oxidoreductase from Clostridium acetobutylicum: a key component of the dioxygen scavenging system in obligatory anaerobes.
Proteins, 78, 2010
|
|
1WUZ
 
 | | Structure of EC1 domain of CNR | | Descriptor: | Pcdha4 protein | | Authors: | Morishita, H, Umitsu, M, Yamaguchi, T, Murata, Y, Shibata, N, Udaka, K, Higuchi, Y, Akutsu, H, Yagi, T, Ikegami, T. | | Deposit date: | 2004-12-09 | | Release date: | 2005-12-13 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structural diversity of the first cadherin domains revealed by the structure of CNR/Protocadherin alpha
To be Published
|
|
1WSP
 
 | | Crystal structure of axin dix domain | | Descriptor: | Axin 1 protein, BENZOIC ACID, MERCURY (II) ION | | Authors: | Shibata, N, Hanamura, T, Yamamoto, R, Ueda, Y, Yamamoto, H, Kikuchi, A, Higuchi, Y. | | Deposit date: | 2004-11-08 | | Release date: | 2006-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of axin dix domain
to be published
|
|
1NY1
 
 | | CRYSTAL STRUCTURE OF B. SUBTILIS POLYSACCHARIDE DEACETYLASE NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET SR127. | | Descriptor: | Probable polysaccharide deacetylase pdaA | | Authors: | Forouhar, F, Edstrom, W, Khan, J, Ma, L, Chiang, Y, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-02-11 | | Release date: | 2003-03-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Polysaccharide Deacetylase (PDAA_BACSU) from B. Subtilis (Pdaa_Bacsu) Northeast Structural Genomics Research Consortium (Nesg) Target Sr127
To be Published
|
|
1NI7
 
 | | NORTHEAST STRUCTURAL GENOMIC CONSORTIUM TARGET ER75 | | Descriptor: | Hypothetical protein ygdK | | Authors: | Liu, G, Chiang, Y, Acton, T, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-12-21 | | Release date: | 2003-06-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-quality homology models derived from NMR and X-ray structures of E. coli proteins YgdK and Suf E suggest that all members of the YgdK/Suf E protein family are enhancers of cysteine desulfurases.
Protein Sci., 14, 2005
|
|
1CPQ
 
 | | CYTOCHROME C' FROM RHODOPSEUDOMONAS CAPSULATA | | Descriptor: | CYTOCHROME C', PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tahirov, T.H, Misaki, S, Meyer, T.E, Cusanovich, M.A, Higuchi, Y, Yasuoka, N. | | Deposit date: | 1995-08-14 | | Release date: | 1996-12-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | High-resolution crystal structures of two polymorphs of cytochrome c' from the purple phototrophic bacterium rhodobacter capsulatus.
J.Mol.Biol., 259, 1996
|
|
1L7Y
 
 | | Solution NMR Structure of C. elegans Protein ZK652.3. NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET WR41. | | Descriptor: | HYPOTHETICAL PROTEIN ZK652.3 | | Authors: | Cort, J.R, Chiang, Y, Zheng, D, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-03-18 | | Release date: | 2002-08-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of conserved eukaryotic protein ZK652.3 from C. elegans: a ubiquitin-like fold.
Proteins, 48, 2002
|
|
3AQX
 
 | | Crystal structure of Bombyx mori beta-GRP/GNBP3 N-terminal domain with laminarihexaoses | | Descriptor: | Beta-1,3-glucan-binding protein, GLYCEROL, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Kanagawa, M, Satoh, T, Ikeda, A, Adachi, Y, Ohno, N, Yamaguchi, Y. | | Deposit date: | 2010-11-22 | | Release date: | 2011-06-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insights into recognition of triple-helical beta-glucans by an insect fungal receptor
J.Biol.Chem., 286, 2011
|
|
7WGQ
 
 | | X-ray structure of human PPAR gamma ligand binding domain-pemafibrate co-crystals obtained by co-crystallization | | Descriptor: | (2~{R})-2-[3-[[1,3-benzoxazol-2-yl-[3-(4-methoxyphenoxy)propyl]amino]methyl]phenoxy]butanoic acid, 15-meric peptide from Nuclear receptor coactivator 1, Isoform 1 of Peroxisome proliferator-activated receptor gamma | | Authors: | Kamata, S, Honda, A, Akahane, M, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Oyama, T, Ishii, I. | | Deposit date: | 2021-12-28 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Functional and Structural Insights into Human PPAR alpha / delta / gamma Subtype Selectivity of Bezafibrate, Fenofibric Acid, and Pemafibrate.
Int J Mol Sci, 23, 2022
|
|
7WGO
 
 | | X-ray structure of human PPAR gamma ligand binding domain-bezafibrate co-rystals obtained by co-crystallization | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, 2-[P-[2-P-CHLOROBENZAMIDO)ETHYL]PHENOXY]-2-METHYLPROPIONIC ACID, Isoform 1 of Peroxisome proliferator-activated receptor gamma | | Authors: | Kamata, S, Honda, A, Akahane, M, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Oyama, T, Ishii, I. | | Deposit date: | 2021-12-28 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Functional and Structural Insights into Human PPAR alpha / delta / gamma Subtype Selectivity of Bezafibrate, Fenofibric Acid, and Pemafibrate.
Int J Mol Sci, 23, 2022
|
|
7WGN
 
 | | X-ray structure of human PPAR delta ligand binding domain-pemafibrate co-crystals obtained by co-crystallization | | Descriptor: | (2~{R})-2-[3-[[1,3-benzoxazol-2-yl-[3-(4-methoxyphenoxy)propyl]amino]methyl]phenoxy]butanoic acid, Peroxisome proliferator-activated receptor delta, octyl beta-D-glucopyranoside | | Authors: | Kamata, S, Honda, A, Akahane, M, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Oyama, T, Ishii, I. | | Deposit date: | 2021-12-28 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.813 Å) | | Cite: | Functional and Structural Insights into Human PPAR alpha / delta / gamma Subtype Selectivity of Bezafibrate, Fenofibric Acid, and Pemafibrate.
Int J Mol Sci, 23, 2022
|
|
7WGP
 
 | | X-ray structure of human PPAR gamma ligand binding domain-fenofibric acid co-crystals obtained by co-crystallization | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, 2-[4-(4-chlorobenzene-1-carbonyl)phenoxy]-2-methylpropanoic acid, Isoform 1 of Peroxisome proliferator-activated receptor gamma | | Authors: | Kamata, S, Honda, A, Akahane, M, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Oyama, T, Ishii, I. | | Deposit date: | 2021-12-28 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Functional and Structural Insights into Human PPAR alpha / delta / gamma Subtype Selectivity of Bezafibrate, Fenofibric Acid, and Pemafibrate.
Int J Mol Sci, 23, 2022
|
|
7WGL
 
 | | X-ray structure of human PPAR delta ligand binding domain-bezafibrate co-crystals obtained by co-crystallization | | Descriptor: | 2-[P-[2-P-CHLOROBENZAMIDO)ETHYL]PHENOXY]-2-METHYLPROPIONIC ACID, Peroxisome proliferator-activated receptor delta, octyl beta-D-glucopyranoside | | Authors: | Kamata, S, Honda, A, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Oyama, T, Ishii, I. | | Deposit date: | 2021-12-28 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | Functional and Structural Insights into Human PPAR alpha / delta / gamma Subtype Selectivity of Bezafibrate, Fenofibric Acid, and Pemafibrate.
Int J Mol Sci, 23, 2022
|
|
6GHP
 
 | | 14-3-3sigma in complex with a TASK3 peptide stabilized by semi-synthetic natural product FC-NAc | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, Potassium channel subfamily K member 9, ... | | Authors: | Andrei, S.A, de Vink, P.J, Brunsveld, L, Ottmann, C, Higuchi, Y. | | Deposit date: | 2018-05-08 | | Release date: | 2018-08-01 | | Last modified: | 2018-10-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Rationally Designed Semisynthetic Natural Product Analogues for Stabilization of 14-3-3 Protein-Protein Interactions.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
3NBS
 
 | | Crystal structure of dimeric cytochrome c from horse heart | | Descriptor: | Cytochrome c, DI(HYDROXYETHYL)ETHER, HEME C, ... | | Authors: | Taketa, M, Komori, H, Hirota, S, Higuchi, Y. | | Deposit date: | 2010-06-04 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cytochrome c polymerization by successive domain swapping at the C-terminal helix
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
