8WM6
 
 | | The structure of PSI-CAC(L-14)of R.salina at 2.7 angstroms resolution | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-3,5,7,9,11,13,15-heptaen-1,17-diynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-(2,6,6-trimethylcyclohexen-1-yl)octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Zhang, S.M, Si, L, Li, M. | | Deposit date: | 2023-10-03 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Growth phase-dependent reorganization of cryptophyte photosystem I antennae.
Commun Biol, 7, 2024
|
|
8D0Z
 
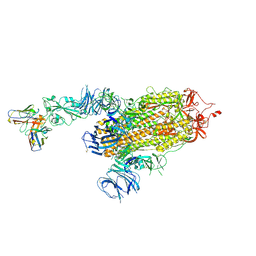 | | S728-1157 IgG in complex with SARS-CoV-2-6P-Mut7 Spike protein (focused refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S728-1157 Fab heavy chain variable region, S728-1157 Fab light chain variable region, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2022-05-26 | | Release date: | 2023-03-22 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Site of vulnerability on SARS-CoV-2 spike induces broadly protective antibody against antigenically distinct Omicron subvariants.
J.Clin.Invest., 133, 2023
|
|
3K5F
 
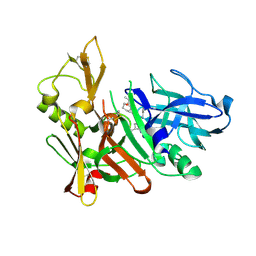 | | Human BACE-1 COMPLEX WITH AYH011 | | Descriptor: | (1R,3S)-3-[1-(acetylamino)-1-methylethyl]-N-[(1S,2S,4R)-1-benzyl-5-(butylamino)-2-hydroxy-4-methyl-5-oxopentyl]cyclohexanecarboxamide, Beta-secretase 1 | | Authors: | Rondeau, J.-M. | | Deposit date: | 2009-10-07 | | Release date: | 2010-05-05 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-based design and synthesis of novel P2/P3 modified, non-peptidic beta-secretase (BACE-1) inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
7FJH
 
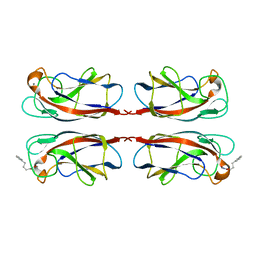 | | LecA from Pseudomonas aeruginosa in complex with 4-Phenylbutyryl hydroxamic acid (CAS: 32153-46-1) | | Descriptor: | CALCIUM ION, N-oxidanyl-4-phenyl-butanamide, PA-I galactophilic lectin | | Authors: | Shanina, S, Kuhaudomlarp, S, Siebs, E, Fuchsberger, F, Denis, M, da Silva Figueiredo Celstino Gomes, P, Clausen, M.H, Seeberger, P.H, Rognan, D, Titz, A, Imberty, A, Rademacher, C. | | Deposit date: | 2021-08-04 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Targeting undruggable carbohydrate recognition sites through focused fragment library design.
Commun Chem, 5, 2022
|
|
7X28
 
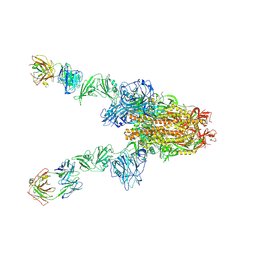 | | MERS-CoV spike complex with S41 neutralizing antibody Fab Class3 (2u1d RBD with 2Fab) | | Descriptor: | Spike glycoprotein, antibody S41 heavy chain, antibody S41 light chain | | Authors: | Zeng, J.W, Zhang, S.Y, Zhou, H.X, Wang, X.W. | | Deposit date: | 2022-02-25 | | Release date: | 2023-01-18 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryoelectron microscopy structures of a human neutralizing antibody bound to MERS-CoV spike glycoprotein.
Front Microbiol, 13, 2022
|
|
7BXU
 
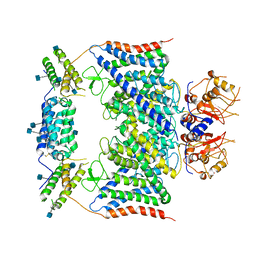 | | CLC-7/Ostm1 membrane protein complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, H(+)/Cl(-) exchange transporter 7, Osteopetrosis-associated transmembrane protein 1 | | Authors: | Zhang, S.S, Yang, M.J. | | Deposit date: | 2020-04-20 | | Release date: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular insights into the human CLC-7/Ostm1 transporter.
Sci Adv, 6, 2020
|
|
7X2A
 
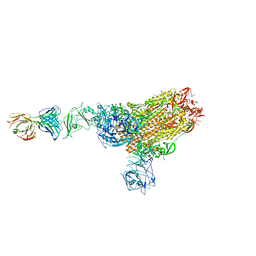 | | MERS-CoV spike complex with S41 neutralizing antibody Fab Class1 (1u2d RBD with 1Fab) | | Descriptor: | MERS-CoV Spike glycoprotein, antibody S41 heavy chain, antibody S41 light chain | | Authors: | Zeng, J.W, Zhang, S.Y, Zhou, H.X, Wang, X.W. | | Deposit date: | 2022-02-25 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryoelectron microscopy structures of a human neutralizing antibody bound to MERS-CoV spike glycoprotein.
Front Microbiol, 13, 2022
|
|
7X26
 
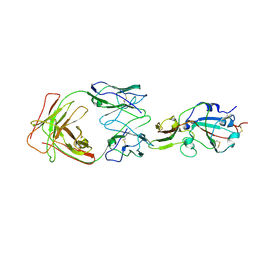 | | S41 neutralizing antibody Fab(MERS-CoV) | | Descriptor: | Spike glycoprotein, antibody S41 heavy chain, antibody S41 light chain | | Authors: | Zeng, J.W, Zhang, S.Y, Wang, X.W. | | Deposit date: | 2022-02-25 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.685 Å) | | Cite: | Cryoelectron microscopy structures of a human neutralizing antibody bound to MERS-CoV spike glycoprotein.
Front Microbiol, 13, 2022
|
|
7X29
 
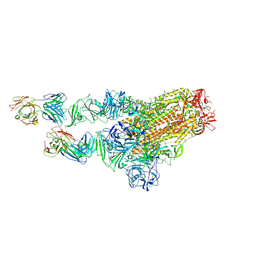 | | MERS-CoV spike complex with S41 neutralizing antibody Fab Class2 (1u2d RBD with 2Fab) | | Descriptor: | Spike glycoprotein, antibody S41 heavy chain, antibody S41 light chain | | Authors: | Zeng, J.W, Zhang, S.Y, Zhou, H.X, Wang, X.W. | | Deposit date: | 2022-02-25 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryoelectron microscopy structures of a human neutralizing antibody bound to MERS-CoV spike glycoprotein.
Front Microbiol, 13, 2022
|
|
7KPF
 
 | | NME2 bound to myristoyl-CoA | | Descriptor: | GLYCEROL, MAGNESIUM ION, Nucleoside diphosphate kinase B, ... | | Authors: | Price, I.R, Lin, H. | | Deposit date: | 2020-11-11 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | A Chemical Proteomic Approach reveals the regulation of NME1/2 and cancer metastasis by Long-Chain Fatty Acyl Coenzyme A
To Be Published
|
|
4U2Z
 
 | | X-ray crystal structure of an Sco GlgEI-V279S/1,2,2-trifluromaltose complex | | Descriptor: | Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase 1, alpha-D-glucopyranose-(1-4)-2-deoxy-2,2-difluoro-alpha-D-arabino-hexopyranosyl fluoride | | Authors: | Ronning, D.R, Lindenberger, J.J. | | Deposit date: | 2014-07-18 | | Release date: | 2015-07-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Synthesis of 2-deoxy-2,2-difluoro-alpha-maltosyl fluoride and its X-ray structure in complex with Streptomyces coelicolor GlgEI-V279S.
Org.Biomol.Chem., 13, 2015
|
|
7DA7
 
 | |
7DAS
 
 | |
7DPI
 
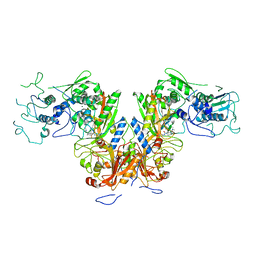 | | Plasmodium falciparum cytoplasmic Phenylalanyl-tRNA synthetase in complex with BRD7929 | | Descriptor: | (8R,9S,10S)-10-[(dimethylamino)methyl]-N-(4-methoxyphenyl)-9-[4-(2-phenylethynyl)phenyl]-1,6-diazabicyclo[6.2.0]decane-6-carboxamide, MAGNESIUM ION, Phenylalanine--tRNA ligase, ... | | Authors: | Manmohan, S, Malhotra, N, Harlos, K, Manickam, Y, Sharma, A. | | Deposit date: | 2020-12-19 | | Release date: | 2022-03-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.597 Å) | | Cite: | Inhibition of Plasmodium falciparum phenylalanine tRNA synthetase provides opportunity for antimalarial drug development.
Structure, 30, 2022
|
|
4LRG
 
 | | Structure of BRD4 bromodomain 1 with a dimethyl thiophene isoxazole azepine carboxamide | | Descriptor: | 2-[(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-[1,2]oxazolo[5,4-c]thieno[2,3-e]azepin-6-yl]acetamide, Bromodomain-containing protein 4 | | Authors: | Ravichandran, S, Jayaram, H, Poy, F, Gehling, V, Hewitt, M, Vaswani, R, Leblanc, Y, Cote, A, Nasveschuk, C, Taylor, A, Harmange, J.-C, Audia, J, Pardo, E, Joshi, S, Sandy, P, Mertz, J, Sims, R, Bergeron, L, Bryant, B, Yellapuntala, S, Nandana, B.S, Birudukota, S, Albrecht, B, Bellon, S. | | Deposit date: | 2013-07-19 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Discovery, Design, and Optimization of Isoxazole Azepine BET Inhibitors.
ACS Med Chem Lett, 4, 2013
|
|
2XL1
 
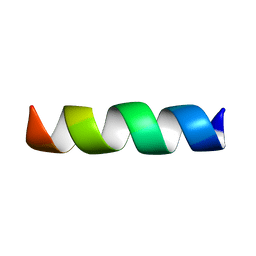 | |
2HVJ
 
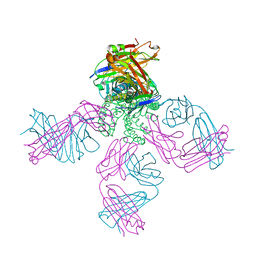 | | Crystal structure of KcsA-Fab-TBA complex in low K+ | | Descriptor: | (2S)-3-HYDROXY-2-(NONANOYLOXY)PROPYL LAURATE, NONAN-1-OL, POTASSIUM ION, ... | | Authors: | Zhou, Y. | | Deposit date: | 2006-07-28 | | Release date: | 2007-02-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystallographic Study of the Tetrabutylammonium Block to the KcsA K(+) Channel.
J.Mol.Biol., 366, 2007
|
|
2HVK
 
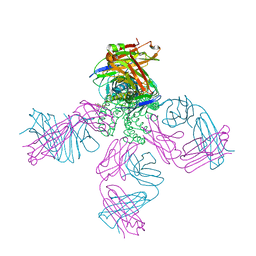 | | crystal structure of the KcsA-Fab-TBA complex in high K+ | | Descriptor: | (2S)-3-HYDROXY-2-(NONANOYLOXY)PROPYL LAURATE, Antibody Fab heavy chain, Antibody Fab light chain, ... | | Authors: | Zhou, Y. | | Deposit date: | 2006-07-28 | | Release date: | 2007-02-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic Study of the Tetrabutylammonium Block to the KcsA K(+) Channel.
J.Mol.Biol., 366, 2007
|
|
2F3E
 
 | | Crystal Structure of the Bace complex with AXQ093, a macrocyclic inhibitor | | Descriptor: | Beta-secretase 1, {(E)-(3R,6S,9R)-3-[(1S,3R)-3-((S)-1 -BUTYLCARBAMOYL-2-METHYL-PROPYLCARB AMOYL)-1-HYDROXY-BUTYL]-6-METHYL-5, 8-DIOXO-1,11-DITHIA-4,7-DIAZA-CYCLO PENTADEC-13-EN-9-YL}-CARBAMIC ACID TERT-BUTYL ESTER | | Authors: | Rondeau, J.-M. | | Deposit date: | 2005-11-21 | | Release date: | 2006-09-05 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure-based design and synthesis of macroheterocyclic peptidomimetic inhibitors of the aspartic protease beta-site amyloid precursor protein cleaving enzyme (BACE).
J.Med.Chem., 49, 2006
|
|
2JRK
 
 | | NMR Structure and Epitope Mapping of Blo t 5 | | Descriptor: | Mite allergen Blo t 5 | | Authors: | Chan, S, Mok, Y. | | Deposit date: | 2007-06-27 | | Release date: | 2008-07-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure and IgE epitopes of Blo t 5, a major dust mite allergen
J.Immunol., 181, 2008
|
|
2F3F
 
 | | Crystal Structure of the Bace complex with BDF488, a macrocyclic inhibitor | | Descriptor: | (2R,4S)-N-BUTYL-4-HYDROXY-2-METHYL- 4-((E)-(4AS,12R,15S,17AS)-15-METHYL -14,17-DIOXO-2,3,4,4A,6,9,11,12,13, 14,15,16,17,17A-TETRADECAHYDRO-1H-5 ,10-DITHIA-1,13,16-TRIAZA-BENZOCYCL OPENTADECEN-12-YL)-BUTYRAMIDE, Beta-secretase 1 | | Authors: | Rondeau, J.-M. | | Deposit date: | 2005-11-21 | | Release date: | 2006-09-05 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based design and synthesis of macroheterocyclic peptidomimetic inhibitors of the aspartic protease beta-site amyloid precursor protein cleaving enzyme (BACE).
J.Med.Chem., 49, 2006
|
|
2GDE
 
 | | Thrombin in complex with inhibitor | | Descriptor: | (R)-3-((2S,3R)-1-((2S,3AR,5S,6S,7AS)-2-(2-(1-CARBAMIMIDOYL-2,5-DIHYDRO-1H-PYRROL-3-YL)ETHYLCARBAMOYL)-5,6-DIHYDROXYOCTAHYDRO-1H-INDOL-1-YL)-3-CHLORO-4-METHYL-1-OXOPENTAN-2-YLAMINO)-2-METHOXY-3-OXOPROPYL HYDROGEN SULFATE, Hirudin, SODIUM ION, ... | | Authors: | Xue, Y. | | Deposit date: | 2006-03-16 | | Release date: | 2007-03-20 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Total synthesis and structural confirmation of chlorodysinosin A.
J.Am.Chem.Soc., 128, 2006
|
|
2BDY
 
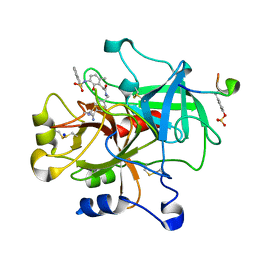 | | thrombin in complex with inhibitor | | Descriptor: | Hirudin IIIB', N-(4-CARBAMIMIDOYL-BENZYL)-2-[2-HYDROXY-6-METHYL-3-(NAPHTHALENE-1-SULFONYLAMINO)-PHENYL]-ACETAMIDE, SODIUM ION, ... | | Authors: | Xue, Y. | | Deposit date: | 2005-10-21 | | Release date: | 2006-10-24 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Phenolic P2/P3 core motif as thrombin inhibitors--design, synthesis, and X-ray co-crystal structure.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
4MJV
 
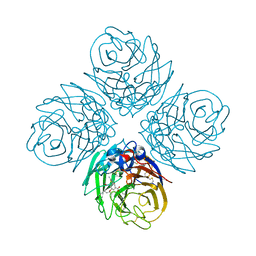 | | Influenza Neuraminidase in complex with a novel antiviral compound | | Descriptor: | (2E,5S,9R,10S)-10-(acetylamino)-2-imino-4-oxo-9-(pentan-3-yloxy)-1-thia-3-azaspiro[4.5]dec-6-ene-7-carboxylic acid, CALCIUM ION, Neuraminidase | | Authors: | Kerry, P.S. | | Deposit date: | 2013-09-04 | | Release date: | 2014-01-15 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Serendipitous discovery of a potent influenza virus a neuraminidase inhibitor.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4MJU
 
 | | Influenza Neuraminidase in complex with a novel antiviral compound | | Descriptor: | (5R,9R,10S)-10-(acetylamino)-2-amino-4-oxo-9-(pentan-3-yloxy)-1-thia-3-azaspiro[4.5]deca-2,6-diene-7-carboxylic acid, CALCIUM ION, Neuraminidase | | Authors: | Kerry, P.S. | | Deposit date: | 2013-09-04 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Serendipitous discovery of a potent influenza virus a neuraminidase inhibitor.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
