1BXN
 
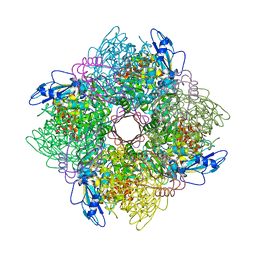 | | THE CRYSTAL STRUCTURE OF RUBISCO FROM ALCALIGENES EUTROPHUS TO 2.7 ANGSTROMS. | | Descriptor: | PHOSPHATE ION, PROTEIN (RIBULOSE BISPHOSPHATE CARBOXYLASE LARGE CHAIN), PROTEIN (RIBULOSE BISPHOSPHATE CARBOXYLASE SMALL CHAIN) | | Authors: | Hansen, S, Vollan, V.B, Hough, E, Andersen, K. | | Deposit date: | 1998-10-06 | | Release date: | 1999-10-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of rubisco from Alcaligenes eutrophus reveals a novel central eight-stranded beta-barrel formed by beta-strands from four subunits.
J.Mol.Biol., 288, 1999
|
|
5H4D
 
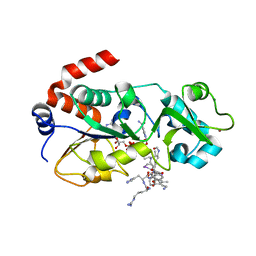 | | Crystal structure of hSIRT3 in complex with a specific agonist Amiodarone hydrochloride | | Descriptor: | (2-butyl-1-benzofuran-3-yl){4-[2-(diethylamino)ethoxy]-3,5-diiodophenyl}methanone, 7-AMINO-4-METHYL-CHROMEN-2-ONE, ARG-HIS-LYS, ... | | Authors: | Zhang, S, Fu, L, Liu, J, Liu, B. | | Deposit date: | 2016-10-31 | | Release date: | 2017-11-08 | | Last modified: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Crystal structure of hSIRT3 in complex with a specific agonist Amiodarone hydrochloride
To Be Published
|
|
5IOO
 
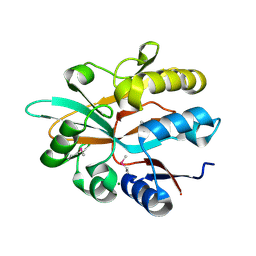 | | Accommodation of massive sequence variation in Nanoarchaeota by the C-type lectin fold | | Descriptor: | AvpA, MAGNESIUM ION | | Authors: | Handa, S, Paul, B, Miller, J, Valentine, D, Ghosh, P. | | Deposit date: | 2016-03-08 | | Release date: | 2016-11-09 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.521 Å) | | Cite: | Conservation of the C-type lectin fold for accommodating massive sequence variation in archaeal diversity-generating retroelements.
Bmc Struct.Biol., 16, 2016
|
|
5KW9
 
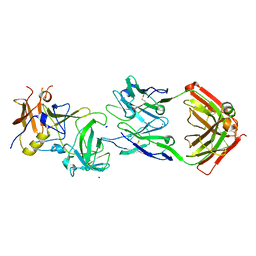 | |
1FEX
 
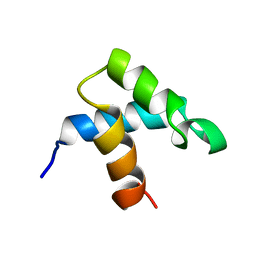 | |
8J64
 
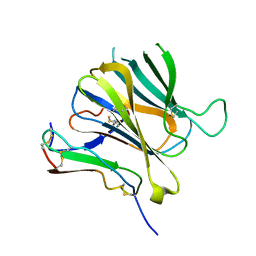 | | Crystal structure of Toxoplasma gondii MIC2-M2AP complex | | Descriptor: | MIC2-associated protein, Micronemal protein MIC2 | | Authors: | Zhang, S, Wang, F.F, Zhang, D.J, Song, G.J, Springer, T.A. | | Deposit date: | 2023-04-24 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into MIC2 recognition by MIC2-associated protein in Toxoplasma gondii.
Commun Biol, 6, 2023
|
|
5JYP
 
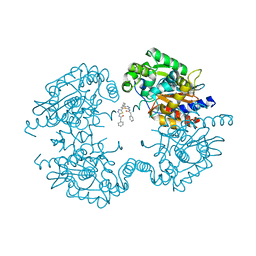 | | Allosteric inhibition of Kidney Isoform of Glutaminase | | Descriptor: | 2-phenyl-~{N}-[5-[(1~{S},3~{S})-3-[5-(2-phenylethanoylamino)-1,3,4-thiadiazol-2-yl]cyclohexyl]-1,3,4-thiadiazol-2-yl]ethanamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Ramachandran, S, Sivaraman, J. | | Deposit date: | 2016-05-15 | | Release date: | 2016-08-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural basis for exploring the allosteric inhibition of human kidney type glutaminase.
Oncotarget, 7, 2016
|
|
1DEG
 
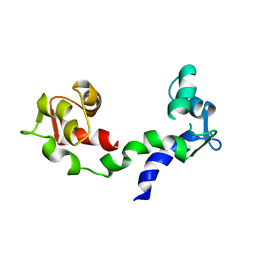 | | THE LINKER OF DES-GLU84 CALMODULIN IS BENT AS SEEN IN THE CRYSTAL STRUCTURE | | Descriptor: | CALCIUM ION, CALMODULIN | | Authors: | Raghunathan, S, Chandross, R, Cheng, B.P, Persechini, A, Sobottk, S.E, Kretsinger, R.H. | | Deposit date: | 1993-06-07 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The linker of des-Glu84-calmodulin is bent.
Proc.Natl.Acad.Sci.Usa, 90, 1993
|
|
6FXB
 
 | | Bovine beta-lactoglobulin variant A at pH 4.0 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Major allergen beta-lactoglobulin, NITRATE ION | | Authors: | Khan, S, Ipsen, R, Almdal, K, Svensson, B, Harris, P. | | Deposit date: | 2018-03-08 | | Release date: | 2018-05-23 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Revealing the Dimeric Crystal and Solution Structure of beta-Lactoglobulin at pH 4 and Its pH and Salt Dependent Monomer-Dimer Equilibrium.
Biomacromolecules, 19, 2018
|
|
1HNA
 
 | | CRYSTAL STRUCTURE OF HUMAN CLASS MU GLUTATHIONE TRANSFERASE GSTM2-2: EFFECTS OF LATTICE PACKING ON CONFORMATIONAL HETEROGENEITY | | Descriptor: | GLUTATHIONE S-(2,4 DINITROBENZENE), GLUTATHIONE S-TRANSFERASE | | Authors: | Raghunathan, S, Chandross, R.J, Kretsinger, R.H, Allison, T.J, Penington, C.J, Rule, G.S. | | Deposit date: | 1993-10-15 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of human class mu glutathione transferase GSTM2-2. Effects of lattice packing on conformational heterogeneity.
J.Mol.Biol., 238, 1994
|
|
5YY5
 
 | | Structural definition of a unique neutralization epitope on the receptor-binding domain of MERS-CoV spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain, Light chain, ... | | Authors: | Zhang, S, Wang, P, Zhou, P, Wang, X, Zhang, L. | | Deposit date: | 2017-12-08 | | Release date: | 2018-08-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Definition of a Unique Neutralization Epitope on the Receptor-Binding Domain of MERS-CoV Spike Glycoprotein
Cell Rep, 24, 2018
|
|
1HNB
 
 | | CRYSTAL STRUCTURE OF HUMAN CLASS MU GLUTATHIONE TRANSFERASE GSTM2-2: EFFECTS OF LATTICE PACKING ON CONFORMATIONAL HETEROGENEITY | | Descriptor: | GLUTATHIONE S-(2,4 DINITROBENZENE), GLUTATHIONE S-TRANSFERASE | | Authors: | Raghunathan, S, Chandross, R.J, Kretsinger, R.H, Allison, T.J, Penington, C.J, Rule, G.S. | | Deposit date: | 1993-10-15 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of human class mu glutathione transferase GSTM2-2. Effects of lattice packing on conformational heterogeneity.
J.Mol.Biol., 238, 1994
|
|
1HNC
 
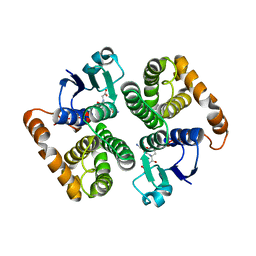 | | CRYSTAL STRUCTURE OF HUMAN CLASS MU GLUTATHIONE TRANSFERASE GSTM2-2: EFFECTS OF LATTICE PACKING ON CONFORMATIONAL HETEROGENEITY | | Descriptor: | GLUTATHIONE S-(2,4 DINITROBENZENE), GLUTATHIONE S-TRANSFERASE | | Authors: | Raghunathan, S, Chandross, R.J, Kretsinger, R.H, Allison, T.J, Penington, C.J, Rule, G.S. | | Deposit date: | 1993-10-15 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human class mu glutathione transferase GSTM2-2. Effects of lattice packing on conformational heterogeneity.
J.Mol.Biol., 238, 1994
|
|
3UGL
 
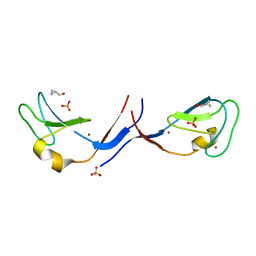 | | Structural and functional characterization of an anesthetic binding site in the second cysteine-rich domain of protein kinase C delta | | Descriptor: | PHOSPHATE ION, Proteine kinase C delta type, ZINC ION, ... | | Authors: | Shanmugasundararaj, S, Stehle, T, Miller, K.W. | | Deposit date: | 2011-11-02 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.357 Å) | | Cite: | Structural and Functional Characterization of an Anesthetic Binding Site in the Second Cysteine-Rich Domain of Protein Kinase Cdelta
Biophys.J., 103, 2012
|
|
3UFF
 
 | |
3UGI
 
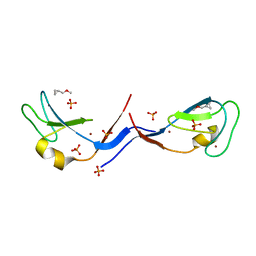 | | Structural and functional characterization of an anesthetic binding site in the second cysteine-rich domain of protein kinase C delta | | Descriptor: | (methoxymethyl)cyclopropane, PHOSPHATE ION, Protein kinase C delta type, ... | | Authors: | Shanmugasundararaj, S, Stehle, T, Miller, K.W. | | Deposit date: | 2011-11-02 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.361 Å) | | Cite: | Structural and Functional Characterization of an Anesthetic Binding Site in the Second Cysteine-Rich Domain of Protein Kinase Cdelta
Biophys.J., 103, 2012
|
|
3UGD
 
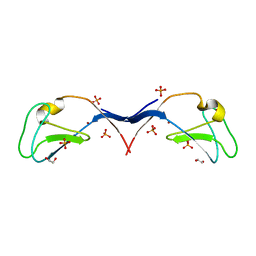 | | Structural and functional characterization of an anesthetic binding site in the second cysteine-rich domain of protein kinase C delta | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, Protein kinase C delta type, ... | | Authors: | Shanmugasundararaj, S, Stehle, T, Miller, K.W. | | Deposit date: | 2011-11-02 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and functional characterization of an anesthetic binding site in the second cysteine-rich domain of protein kinase C delta
Biophys.J., 103, 2012
|
|
6SHH
 
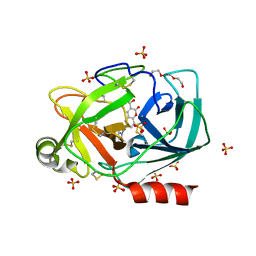 | |
5ZS3
 
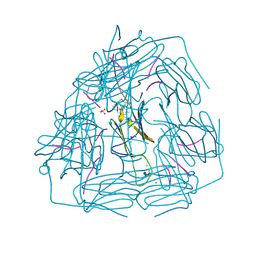 | | Small heat shock protein from M. marinum:Form-1 | | Descriptor: | CHLORIDE ION, GLY-ARG-LEU-LEU-PRO, Molecular chaperone (Small heat shock protein), ... | | Authors: | Bhandari, S, Suguna, K. | | Deposit date: | 2018-04-27 | | Release date: | 2019-01-30 | | Last modified: | 2019-04-17 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Dodecameric structure of a small heat shock protein from Mycobacterium marinum M.
Proteins, 87, 2019
|
|
5ZS6
 
 | |
5ZUL
 
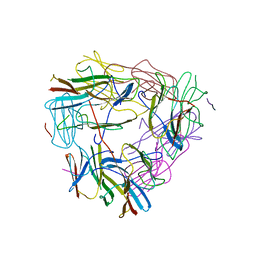 | |
5MA8
 
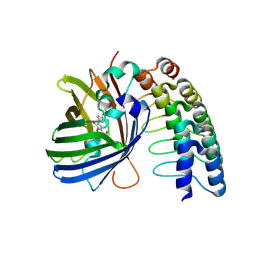 | | GFP-binding DARPin 3G124nc | | Descriptor: | GA-binding protein subunit beta-1, Green fluorescent protein | | Authors: | Hansen, S, Stueber, J, Ernst, P, Koch, A, Bojar, D, Batyuk, A, Plueckthun, A. | | Deposit date: | 2016-11-03 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Design and applications of a clamp for Green Fluorescent Protein with picomolar affinity.
Sci Rep, 7, 2017
|
|
5MFJ
 
 | | Designed armadillo repeat protein YIII(Dq.V2)4CqI in complex with peptide (KR)5 | | Descriptor: | (KR)5, YIII(Dq.V2)4CqI | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
5MFO
 
 | | Designed armadillo repeat protein YIIIM3AIII | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, YIIIM3AIII | | Authors: | Hansen, S, Ernst, P, Reichen, C, Ewald, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Curvature of designed armadillo repeat proteins allows modular peptide binding.
J. Struct. Biol., 201, 2018
|
|
6H42
 
 | | crystal structure of the human TGT catalytic subunit QTRT1 | | Descriptor: | BROMIDE ION, CHLORIDE ION, GLUTAMIC ACID, ... | | Authors: | Johannsson, S, Neumann, P, Ficner, R. | | Deposit date: | 2018-07-20 | | Release date: | 2018-09-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of the Human tRNA Guanine Transglycosylase Catalytic Subunit QTRT1.
Biomolecules, 8, 2018
|
|
