6H34
 
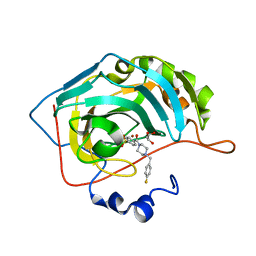 | | The crystal structure of human carbonic anhydrase II in complex with 4-[(4-fluorophenyl)methyl]-1-piperazinyl]benzenesulfonamide. | | Descriptor: | 4-[4-[(4-fluorophenyl)methyl]piperazin-1-yl]carbonylbenzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Buemi, M.R, Di Fiore, A, De Luca, L, Ferro, S, Mancuso, F, Monti, S.M, Buonanno, M, Angeli, A, Russo, E, De Sarro, G, Supuran, C.T, De Simone, G, Gitto, R. | | Deposit date: | 2018-07-17 | | Release date: | 2018-12-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Exploring structural properties of potent human carbonic anhydrase inhibitors bearing a 4-(cycloalkylamino-1-carbonyl)benzenesulfonamide moiety.
Eur J Med Chem, 163, 2018
|
|
6H38
 
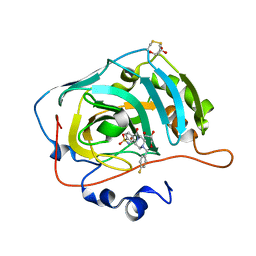 | | The crystal structure of human carbonic anhydrase VII in complex with 4-[(4-fluorophenyl)methyl]-1-piperazinyl]benzenesulfonamide. | | Descriptor: | 4-[4-[(4-fluorophenyl)methyl]piperazin-1-yl]carbonylbenzenesulfonamide, Carbonic anhydrase 7, GLYCEROL, ... | | Authors: | Buemi, M.R, Di Fiore, A, De Luca, L, Ferro, S, Mancuso, F, Monti, S.M, Buonanno, M, Angeli, A, Russo, E, De Sarro, G, Supuran, C.T, De Simone, G, Gitto, R. | | Deposit date: | 2018-07-17 | | Release date: | 2018-12-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Exploring structural properties of potent human carbonic anhydrase inhibitors bearing a 4-(cycloalkylamino-1-carbonyl)benzenesulfonamide moiety.
Eur J Med Chem, 163, 2018
|
|
6H36
 
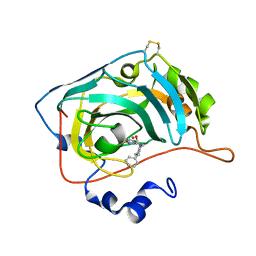 | | The crystal structure of human carbonic anhydrase VII in complex with 4-(4-phenylpiperidine-1-carbonyl)benzenesulfonamide. | | Descriptor: | 4-(4-phenylpiperidin-1-yl)carbonylbenzenesulfonamide, Carbonic anhydrase 7, ZINC ION | | Authors: | Buemi, M.R, Di Fiore, A, De Luca, L, Ferro, S, Mancuso, F, Monti, S.M, Buonanno, M, Angeli, A, Russo, E, De Sarro, G, Supuran, C.T, De Simone, G, Gitto, R. | | Deposit date: | 2018-07-17 | | Release date: | 2018-12-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Exploring structural properties of potent human carbonic anhydrase inhibitors bearing a 4-(cycloalkylamino-1-carbonyl)benzenesulfonamide moiety.
Eur J Med Chem, 163, 2018
|
|
6RG3
 
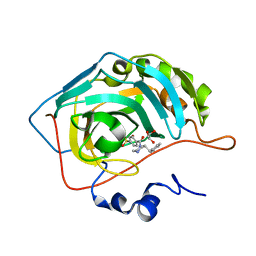 | | Crystal structure of human Carbonic anhydrase II in complex with (R)-4-(2-benzylpiperazin-1-yl)benzenesulfonamide | | Descriptor: | 4-[(2~{R})-2-(phenylmethyl)piperazin-1-yl]carbonylbenzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Ferraroni, M, Angeli, A, Supuran, C. | | Deposit date: | 2019-04-16 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Sulfonamides incorporating piperazine bioisosteres as potent human carbonic anhydrase I, II, IV and IX inhibitors.
Bioorg.Chem., 91, 2019
|
|
6RG4
 
 | | Crystal structure of human Carbonic anhydrase II in complex with (R)-4-(2-benzyl-4-methylpiperazin-1-yl)benzenesulfonamide | | Descriptor: | 4-[(3~{S})-4-methyl-3-(phenylmethyl)piperazin-1-yl]carbonylbenzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Ferraroni, M, Angeli, A, Supuran, C. | | Deposit date: | 2019-04-16 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Sulfonamides incorporating piperazine bioisosteres as potent human carbonic anhydrase I, II, IV and IX inhibitors.
Bioorg.Chem., 91, 2019
|
|
6RHJ
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 4-(4-benzyl-1,4-diazepane-1-carbonyl)benzenesulfonamide | | Descriptor: | 4-[[4-(phenylmethyl)-1,4-diazepan-1-yl]carbonyl]benzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Ferraroni, M, Angeli, A, Supuran, C.T. | | Deposit date: | 2019-04-21 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Sulfonamides incorporating piperazine bioisosteres as potent human carbonic anhydrase I, II, IV and IX inhibitors.
Bioorg.Chem., 91, 2019
|
|
6XZS
 
 | | Crystal structure of human carbonic anhydrase I in complex with 4-(3-(2-((4-fluorobenzyl)amino)ethyl)ureido) benzenesulfonamide | | Descriptor: | 1-[2-[(4-fluorophenyl)methylamino]ethyl]-3-(4-sulfamoylphenyl)urea, Carbonic anhydrase 1, ZINC ION | | Authors: | Zanotti, G, Majid, A, Bozdag, M, Angeli, A, Carta, F, Berto, F, Supuran, C.T. | | Deposit date: | 2020-02-05 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Benzylaminoethyureido-Tailed Benzenesulfonamides: Design, Synthesis, Kinetic and X-ray Investigations on Human Carbonic Anhydrases.
Int J Mol Sci, 21, 2020
|
|
6XZY
 
 | | crystal structure of human carbonic anhydrase I in complex with 4-(3-(2-((2-fluorobenzyl)amino)ethyl)ureido) benzenesulfonamide | | Descriptor: | 1-[2-[(2-fluorophenyl)methylamino]ethyl]-3-(3-sulfamoylphenyl)urea, Carbonic anhydrase 1, ZINC ION | | Authors: | Zanotti, G, Majid, A, Bozdag, M, Angeli, A, Carta, F, Berto, P, Supuran, C. | | Deposit date: | 2020-02-05 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Benzylaminoethyureido-Tailed Benzenesulfonamides: Design, Synthesis, Kinetic and X-ray Investigations on Human Carbonic Anhydrases.
Int J Mol Sci, 21, 2020
|
|
6XZX
 
 | | crystal structure of human carbonic anhydrase I in complex with 4-(3-(2-((2-fluorobenzyl)amino)ethyl)ureido) benzenesulfonamide | | Descriptor: | 1-[2-[(phenylmethyl)amino]ethyl]-3-(3-sulfamoylphenyl)urea, Carbonic anhydrase 1, ZINC ION | | Authors: | Zanotti, G, Majid, A, Bozdag, M, Angeli, A, Carta, F, Berto, P, Supuran, C. | | Deposit date: | 2020-02-05 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Benzylaminoethyureido-Tailed Benzenesulfonamides: Design, Synthesis, Kinetic and X-ray Investigations on Human Carbonic Anhydrases.
Int J Mol Sci, 21, 2020
|
|
6Y00
 
 | | crystal structure of human carbonic anhydrase I in complex with 4-(3-(2-((2-fluorobenzyl)amino)ethyl)ureido) benzenesulfonamide | | Descriptor: | 1-[2-[(4-bromanyl-2-oxidanyl-phenyl)methylamino]ethyl]-3-(3-sulfamoylphenyl)urea, Carbonic anhydrase 1, ZINC ION | | Authors: | Zanotti, G, Majid, A, Bozdag, M, Angeli, A, Carta, F, Berto, P, Supuran, C. | | Deposit date: | 2020-02-05 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Benzylaminoethyureido-Tailed Benzenesulfonamides: Design, Synthesis, Kinetic and X-ray Investigations on Human Carbonic Anhydrases.
Int J Mol Sci, 21, 2020
|
|
6XZO
 
 | | Crystal structure of human carbonic anhydrase I in complex with 4-(3-(2-((4-bromo-2-hydroxybenzyl)amino)ethyl)ureido) benzenesulfonamide | | Descriptor: | 1-[2-[(4-bromanyl-2-oxidanyl-phenyl)methylamino]ethyl]-3-(4-sulfamoylphenyl)urea, Carbonic anhydrase 1, ZINC ION | | Authors: | Zanotti, G, Majid, A, Bozdag, M, Angeli, A, Carta, F, Berto, P, Supuran, C. | | Deposit date: | 2020-02-05 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Benzylaminoethyureido-Tailed Benzenesulfonamides: Design, Synthesis, Kinetic and X-ray Investigations on Human Carbonic Anhydrases.
Int J Mol Sci, 21, 2020
|
|
7Z57
 
 | | Crystal structure of Human Serum Albumin in complex with surfactant GenX (2,3,3,3-tetrafluoro-2-(heptafluoropropoxy) propanoate) | | Descriptor: | (2R)-2,3,3,3-tetrakis(fluoranyl)-2-[1,1,2,2,3,3,3-heptakis(fluoranyl)propoxy]propanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Albumin, ... | | Authors: | Liberi, S, Moro, G, Vascon, F, Linciano, S, De Toni, L, Angelin, A, Cendron, L. | | Deposit date: | 2022-03-08 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Investigation of the Interaction between Human Serum Albumin and Branched Short-Chain Perfluoroalkyl Compounds.
Chem.Res.Toxicol., 35, 2022
|
|
3CWY
 
 | | Structure of CagD from H. pylori pathogenicity island crystallized in the presence of Cu(II) ions | | Descriptor: | COPPER (II) ION, protein CagD | | Authors: | Cendron, L, Zanotti, G, Angelini, A, Barison, N, Couturier, M, Stein, M. | | Deposit date: | 2008-04-23 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Helicobacter pylori CagD (HP0545, Cag24) protein is essential for CagA translocation and maximal induction of interleukin-8 secretion.
J.Mol.Biol., 386, 2009
|
|
3CWX
 
 | | Crystal structure of cagd from helicobacter pylori pathogenicity island | | Descriptor: | protein CagD | | Authors: | Cendron, L, Zanotti, G, Angelini, A, Barison, N, Couturier, M, Stein, M. | | Deposit date: | 2008-04-23 | | Release date: | 2008-12-30 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Helicobacter pylori CagD (HP0545, Cag24) protein is essential for CagA translocation and maximal induction of interleukin-8 secretion.
J.Mol.Biol., 386, 2009
|
|
8PDE
 
 | | Crystal Structure of the MADS-box/MEF2 Domain of MEF2D bound to dsDNA and HDAC4 deacetylase binding motif | | Descriptor: | DNA (5'-D(P*AP*AP*CP*TP*AP*TP*TP*TP*AP*TP*AP*AP*GP*A)-3'), DNA (5'-D(P*TP*CP*TP*TP*AP*TP*AP*AP*AP*TP*AP*GP*TP*T)-3'), HDAC4 (histone deacetylase 4) binding motif peptide:GSGEVKMKLQEFVLNKK, ... | | Authors: | Chinellato, M, Carli, A, Perin, S, Mazzocato, Y, Biondi, B, Di Giorgio, E, Brancolini, C, Angelini, A, Cendron, L. | | Deposit date: | 2023-06-12 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Folding of Class IIa HDAC Derived Peptides into alpha-helices Upon Binding to Myocyte Enhancer Factor-2 in Complex with DNA.
J.Mol.Biol., 436, 2024
|
|
1S2X
 
 | | Crystal structure of Cag-Z from Helicobacter pylori | | Descriptor: | Cag-Z, ISOPROPYL ALCOHOL | | Authors: | Cendron, L, Seydel, A, Angelini, A, Battistutta, R, Zanotti, G. | | Deposit date: | 2004-01-12 | | Release date: | 2004-07-27 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of CagZ, a protein from the Helicobacter pylori pathogenicity island that encodes for a type IV secretion system
J.Mol.Biol., 340, 2004
|
|
2G3V
 
 | | Crystal structure of CagS (HP0534, Cag13) from Helicobacter pylori | | Descriptor: | (UNK)(UNK)(UNK)(UNK)(UNK)(MSE)(UNK), CAG pathogenicity island protein 13 | | Authors: | Cendron, L, Tasca, E, Angelini, A, Seydel, A, Battistutta, R, Montecucco, C, Zanotti, G. | | Deposit date: | 2006-02-21 | | Release date: | 2007-03-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of CagS from helicobacter pylori
To be Published
|
|
8C84
 
 | | Crystal structure of MADS-box/MEF2D N-terminal domain complex | | Descriptor: | DNA (5'-D(P*AP*AP*CP*TP*AP*TP*TP*TP*AP*TP*AP*AP*GP*A)-3'), DNA (5'-D(P*TP*CP*TP*TP*AP*TP*AP*AP*AP*TP*AP*GP*TP*T)-3'), MEF2D protein | | Authors: | Chinellato, M, Carli, A, Perin, S, Mazzoccato, Y, Di Giorgio, E, Brancolini, C, Angelini, A, Cendron, L. | | Deposit date: | 2023-01-18 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Folding of Class IIa HDAC Derived Peptides into alpha-helices Upon Binding to Myocyte Enhancer Factor-2 in Complex with DNA.
J.Mol.Biol., 436, 2024
|
|
8Q9R
 
 | | Crystal structure of MADS-box/MEF2D N-terminal domain bound to dsDNA and HDAC9 deacetylase binding motif | | Descriptor: | Histone deacetylase 9 (HDAC9) binding motif peptide: EVKQKLQEFLLSKS, MADS box dsDNA: AACTATTTATAAGA, MADS box dsDNA: TCTTATAAATAGT, ... | | Authors: | Chinellato, M, Carli, A, Perin, S, Mazzoccato, Y, Di Giorgio, E, Brancolini, C, Angelini, A, Cendron, L. | | Deposit date: | 2023-08-20 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Folding of Class IIa HDAC Derived Peptides into alpha-helices Upon Binding to Myocyte Enhancer Factor-2 in Complex with DNA.
J.Mol.Biol., 436, 2024
|
|
8Q9Q
 
 | | Crystal Structure of the MADS-box/MEF2 Domain of MEF2D bound to dsDNA and HDAC7 deacetylase binding motif | | Descriptor: | HDAC7 (histone deacetylase 7) binding motif peptide: GLY-VAL-VAL-LYS-GLN-LYS-LEU-ALA-GLU-VAL-ILE-LEU-LYS-LYS-GLN, MADS box dsDNA: AACTATTTATAAGA, MADS box dsDNA: TCTTATAAATAGTT, ... | | Authors: | Chinellato, M, Carli, A, Perin, S, Mazzocato, Y, Biondi, B, Di Giorgio, E, Brancolini, C, Angelini, A, Cendron, L. | | Deposit date: | 2023-08-20 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Folding of Class IIa HDAC Derived Peptides into alpha-helices Upon Binding to Myocyte Enhancer Factor-2 in Complex with DNA.
J.Mol.Biol., 436, 2024
|
|
8Q9N
 
 | | Crystal Structure of the MADS-box/MEF2 Domain of MEF2D bound to dsDNA and MITR deacetylase binding motif mutant L151V. | | Descriptor: | DIMETHYL SULFOXIDE, DNA MADS box, MEF2D protein, ... | | Authors: | Chinellato, M, Carli, A, Perin, S, Mazzocato, Y, Biondi, B, Di Giorgio, E, Brancolini, C, Angelini, A, Cendron, L. | | Deposit date: | 2023-08-20 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Folding of Class IIa HDAC Derived Peptides into alpha-helices Upon Binding to Myocyte Enhancer Factor-2 in Complex with DNA.
J.Mol.Biol., 436, 2024
|
|
2RD3
 
 | |
8Q9P
 
 | | Crystal Structure of the MADS-box/MEF2 Domain of MEF2D bound to dsDNA and HDAC5 deacetylase binding motif | | Descriptor: | HDAC5 (histone deacetylase 5) binding motif peptide: TRP-GLY-SER-GLY-GLU-VAL-LYS-LEU-ARG-LEU-GLN-GLU-PHE-LEU-LEU-SER-LYS-SER, MADS box dsDNA fw: AACTATTTATAAGA, MADS box dsNA rev:TCTTATAAATAGTT, ... | | Authors: | Chinellato, M, Carli, A, Perin, S, Mazzocato, Y, Biondi, B, Di Giorgio, E, Brancolini, C, Angelini, A, Cendron, L. | | Deposit date: | 2023-08-20 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Folding of Class IIa HDAC Derived Peptides into alpha-helices Upon Binding to Myocyte Enhancer Factor-2 in Complex with DNA.
J.Mol.Biol., 436, 2024
|
|
8OI2
 
 | | Crystal structure of Alb1 megabody in complex with human serum albumin | | Descriptor: | Alb1 Megabody, Albumin | | Authors: | De Felice, S, Zoia, G, Romanyuk, Z, Pardon, E, Steyaert, J, Angelini, A, Cendron, L. | | Deposit date: | 2023-03-22 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of human serum albumin in complex with megabody reveals unique human and murine cross-reactive binding site.
Protein Sci., 33, 2024
|
|
7ZRR
 
 | | Crystal structure of human Urokinase-type plasminogen activator in complex with bicycle peptide inhibitor UK965 | | Descriptor: | 1,2-ETHANEDIOL, 1,3,5-tris(bromomethyl)benzene, AMINO GROUP, ... | | Authors: | Caregnato, A, Angela, P, Mazzoccato, Y, Frasson, N, Angelini, A, Cendron, L. | | Deposit date: | 2022-05-05 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of human Urokinase-type plasminogen activator in complex with bicycle peptide inhibitor UK965
To Be Published
|
|
