5E66
 
 | | The complex structure of Hemagglutinin-esterase-fusion mutant protein from the influenza D virus with receptor analog 9-N-Ac-Sia | | Descriptor: | (6R)-5-acetamido-6-[(1S,2S)-3-acetamido-1,2-dihydroxypropyl]-3,5-dideoxy-beta-L-threo-hex-2-ulopyranosonic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Song, H, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2015-10-09 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | An Open Receptor-Binding Cavity of Hemagglutinin-Esterase-Fusion Glycoprotein from Newly-Identified Influenza D Virus: Basis for Its Broad Cell Tropism
PLoS Pathog., 12, 2016
|
|
5E65
 
 | | The complex structure of Hemagglutinin-esterase-fusion mutant protein from the influenza D virus with receptor analog 9-O-Ac-3'SLN (Tr322) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 9-O-acetyl-5-acetamido-3,5-dideoxy-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid-(2-3)-beta-D-galactopyranose, ... | | Authors: | Song, H, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2015-10-09 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | An Open Receptor-Binding Cavity of Hemagglutinin-Esterase-Fusion Glycoprotein from Newly-Identified Influenza D Virus: Basis for Its Broad Cell Tropism
PLoS Pathog., 12, 2016
|
|
5E62
 
 | | HEF-mut with Tr323 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 9-O-acetyl-5-acetamido-3,5-dideoxy-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-azidoethyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, ... | | Authors: | Song, H, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2015-10-09 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | An Open Receptor-Binding Cavity of Hemagglutinin-Esterase-Fusion Glycoprotein from Newly-Identified Influenza D Virus: Basis for Its Broad Cell Tropism
PLoS Pathog., 12, 2016
|
|
5E64
 
 | | Hemagglutinin-esterase-fusion protein structure of influenza D virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CACODYLATE ION, ... | | Authors: | Song, H, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2015-10-09 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An Open Receptor-Binding Cavity of Hemagglutinin-Esterase-Fusion Glycoprotein from Newly-Identified Influenza D Virus: Basis for Its Broad Cell Tropism
PLoS Pathog., 12, 2016
|
|
2X1L
 
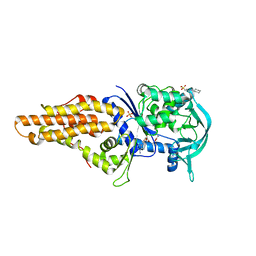 | | Crystal structure of Mycobacterium smegmatis methionyl-tRNA synthetase in complex with methionine and adenosine | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, ADENOSINE, DIHYDROGENPHOSPHATE ION, ... | | Authors: | Ingvarsson, H, Jones, T.A, Unge, T. | | Deposit date: | 2009-12-31 | | Release date: | 2010-07-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Flexibility and Communication within the Structure of the Mycobacterium Smegmatis Methionyl-tRNA Synthetase.
FEBS J., 277, 2010
|
|
2X1M
 
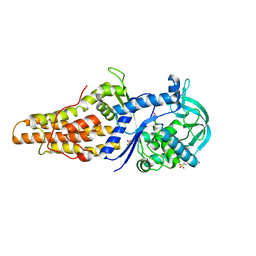 | | Crystal structure of Mycobacterium smegmatis methionyl-tRNA synthetase in complex with methionine | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, DIHYDROGENPHOSPHATE ION, METHIONINE, ... | | Authors: | Ingvarsson, H, Jones, T.A, Unge, T. | | Deposit date: | 2009-12-31 | | Release date: | 2010-07-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Flexibility and Communication within the Structure of the Mycobacterium Smegmatis Methionyl-tRNA Synthetase.
FEBS J., 277, 2010
|
|
8SSW
 
 | |
6JO8
 
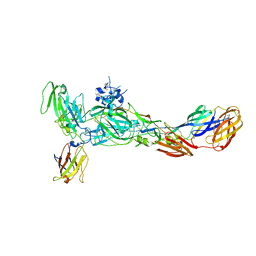 | | The complex structure of CHIKV envelope glycoprotein bound to human MXRA8 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHIKV E1, ... | | Authors: | Song, H, Zhao, Z, Qi, J, Gao, F, Gao, F.G. | | Deposit date: | 2019-03-20 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.495 Å) | | Cite: | Molecular Basis of Arthritogenic Alphavirus Receptor MXRA8 Binding to Chikungunya Virus Envelope Protein.
Cell, 177, 2019
|
|
6O5F
 
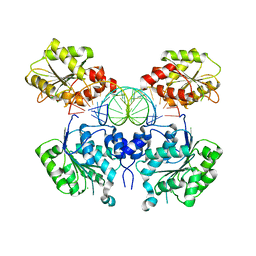 | | Crystal structure of DEAD-box RNA helicase DDX3X at pre-unwound state | | Descriptor: | ATP-dependent RNA helicase DDX3X, CHLORIDE ION, RNA (5'-R(P*CP*AP*AP*GP*GP*UP*CP*AP*UP*UP*CP*GP*CP*AP*AP*GP*AP*GP*UP*GP*GP*CP*C)-3') | | Authors: | Song, H, Ji, X. | | Deposit date: | 2019-03-02 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | The mechanism of RNA duplex recognition and unwinding by DEAD-box helicase DDX3X.
Nat Commun, 10, 2019
|
|
6JO7
 
 | | Crystal structure of mouse MXRA8 | | Descriptor: | Matrix remodeling-associated protein 8 | | Authors: | Song, H, Zhao, Z, Qi, J, Gao, F, Gao, G.F. | | Deposit date: | 2019-03-20 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Basis of Arthritogenic Alphavirus Receptor MXRA8 Binding to Chikungunya Virus Envelope Protein.
Cell, 177, 2019
|
|
8ANO
 
 | | Fucosylated mixed-chirality linear peptide FHP8 bound to the fucose binding lectin LecB PA-IIL from Pseudomonas aeruginosa at 1.3 Angstrom resolution. | | Descriptor: | 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid, CALCIUM ION, Fucose-binding lectin PA-IIL, ... | | Authors: | Personne, H, Stocker, A, Reymond, J.-L. | | Deposit date: | 2022-08-05 | | Release date: | 2023-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | To Fold or Not to Fold: Diastereomeric Optimization of an alpha-Helical Antimicrobial Peptide.
J.Med.Chem., 66, 2023
|
|
8ANR
 
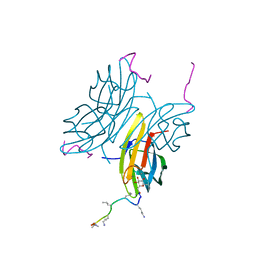 | | Fucosylated alternate chirality linear peptide FHP30 bound to the fucose binding lectin LecB PA-IIL from Pseudomonas aeruginosa at 1.6 Angstrom resolution. | | Descriptor: | 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid, CALCIUM ION, Fucose-binding lectin PA-IIL, ... | | Authors: | Personne, H, Stocker, A, Reymond, J.-L. | | Deposit date: | 2022-08-05 | | Release date: | 2023-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | To Fold or Not to Fold: Diastereomeric Optimization of an alpha-Helical Antimicrobial Peptide.
J.Med.Chem., 66, 2023
|
|
8AOO
 
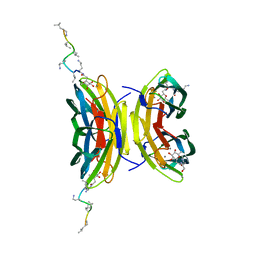 | | Fucosylated mixed-chirality linear peptide FHP31 bound to the fucose binding lectin LecB PA-IIL from Pseudomonas aeruginosa at 1.2 Angstrom resolution. | | Descriptor: | 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid, CALCIUM ION, Fucose-binding lectin PA-IIL, ... | | Authors: | Personne, H, Stocker, A, Reymond, J.-L. | | Deposit date: | 2022-08-08 | | Release date: | 2023-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | To Fold or Not to Fold: Diastereomeric Optimization of an alpha-Helical Antimicrobial Peptide.
J.Med.Chem., 66, 2023
|
|
8AN9
 
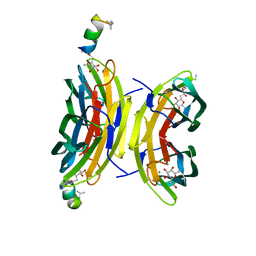 | | Fucosylated mixed-chirality linear peptide FHP5 bound to the fucose binding lectin LecB PA-IIL from Pseudomonas aeruginosa at 1.3 Angstrom resolution. | | Descriptor: | 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid, CALCIUM ION, Fucose-binding lectin PA-IIL, ... | | Authors: | Personne, H, Stocker, A, Reymond, J.-L. | | Deposit date: | 2022-08-04 | | Release date: | 2023-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.273 Å) | | Cite: | To Fold or Not to Fold: Diastereomeric Optimization of an alpha-Helical Antimicrobial Peptide.
J.Med.Chem., 66, 2023
|
|
5IY3
 
 | | Zika Virus Non-structural Protein NS1 | | Descriptor: | Genome polyprotein | | Authors: | Song, H, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2016-03-23 | | Release date: | 2016-04-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Zika virus NS1 structure reveals diversity of electrostatic surfaces among flaviviruses
Nat.Struct.Mol.Biol., 23, 2016
|
|
1FQ1
 
 | | CRYSTAL STRUCTURE OF KINASE ASSOCIATED PHOSPHATASE (KAP) IN COMPLEX WITH PHOSPHO-CDK2 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CELL DIVISION PROTEIN KINASE 2, CYCLIN-DEPENDENT KINASE INHIBITOR 3, ... | | Authors: | Song, H, Hanlon, N, Brown, N.R, Noble, M.E.M, Johnson, L.N, Barford, D. | | Deposit date: | 2000-09-01 | | Release date: | 2001-05-09 | | Last modified: | 2018-03-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Phosphoprotein-protein interactions revealed by the crystal structure of kinase-associated phosphatase in complex with phosphoCDK2.
Mol.Cell, 7, 2001
|
|
1FPZ
 
 | | CRYSTAL STRUCTURE ANALYSIS OF KINASE ASSOCIATED PHOSPHATASE (KAP) WITH A SUBSTITUTION OF THE CATALYTIC SITE CYSTEINE (CYS140) TO A SERINE | | Descriptor: | CYCLIN-DEPENDENT KINASE INHIBITOR 3, SULFATE ION | | Authors: | Song, H, Hanlon, N, Brown, N.R, Noble, M.E.M, Johnson, L.N, Barford, D. | | Deposit date: | 2000-09-01 | | Release date: | 2001-05-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphoprotein-protein interactions revealed by the crystal structure of kinase-associated phosphatase in complex with phosphoCDK2.
Mol.Cell, 7, 2001
|
|
403D
 
 | | 5'-D(*CP*GP*CP*(HYD)AP*AP*AP*TP*TP*TP*GP*CP*G)-3', 2'-(4-ETHOXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE | | Descriptor: | 2'-(4-ETHOXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*(IGU)P*AP*AP*TP*TP*TP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Robinson, H, Gao, Y.-G, Bauer, C, Roberts, C, Switzer, C, Wang, A.H.-J. | | Deposit date: | 1998-06-10 | | Release date: | 1998-07-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | 2'-Deoxyisoguanosine adopts more than one tautomer to form base pairs with thymidine observed by high-resolution crystal structure analysis.
Biochemistry, 37, 1998
|
|
5T16
 
 | |
5OUF
 
 | |
1OOW
 
 | | The crystal structure of the spinach plastocyanin double mutant G8D/L12E gives insight into its low reactivity towards photosystem 1 and cytochrome f | | Descriptor: | COPPER (II) ION, Plastocyanin, chloroplast | | Authors: | Jansson, H, Okvist, M, Jacobson, F, Ejdeback, M, Hansson, O, Sjolin, L. | | Deposit date: | 2003-03-04 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the spinach plastocyanin double mutant G8D/L12E gives insight into its low reactivity towards photosystem 1 and cytochrome f.
Biochim.Biophys.Acta, 1607, 2003
|
|
1Q9C
 
 | |
1XD4
 
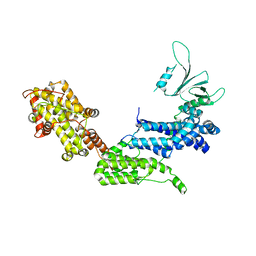 | | Crystal structure of the DH-PH-cat module of Son of Sevenless (SOS) | | Descriptor: | Son of sevenless protein homolog 1 | | Authors: | Sondermann, H, Soisson, S.M, Boykevisch, S, Yang, S.S, Bar-Sagi, D, Kuriyan, J. | | Deposit date: | 2004-09-03 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.64 Å) | | Cite: | Structural analysis of autoinhibition in the ras activator son of sevenless.
Cell(Cambridge,Mass.), 119, 2004
|
|
1XD2
 
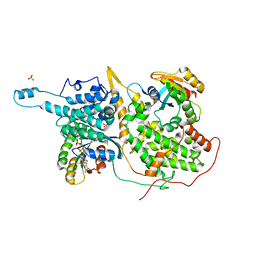 | | Crystal Structure of a ternary Ras:SOS:Ras*GDP complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Sondermann, H, Soisson, S.M, Boykevisch, S, Yang, S.S, Bar-Sagi, D, Kuriyan, J. | | Deposit date: | 2004-09-03 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of autoinhibition in the ras activator son of sevenless.
Cell(Cambridge,Mass.), 119, 2004
|
|
1XDV
 
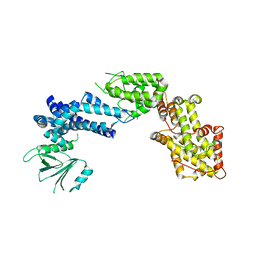 | | Experimentally Phased Structure of Human the Son of Sevenless protein at 4.1 Ang. | | Descriptor: | Son of sevenless protein homolog 1 | | Authors: | Sondermann, H, Soisson, S.M, Boykevisch, S, Yang, S.S, Bar-Sagi, D, Kuriyan, J. | | Deposit date: | 2004-09-08 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structural analysis of autoinhibition in the ras activator son of sevenless.
Cell(Cambridge,Mass.), 119, 2004
|
|
