6ABL
 
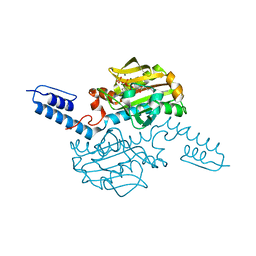 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with oBrZLys | | Descriptor: | (2S)-2-azanyl-6-[(2-bromophenyl)methoxycarbonylamino]hexanoic acid, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-22 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
6ABM
 
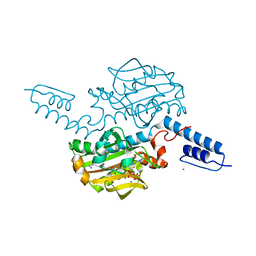 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with pTmdZLys | | Descriptor: | (2S)-2-azanyl-6-[[4-[3-(trifluoromethyl)-1,2-diazirin-3-yl]phenyl]methoxycarbonylamino]hexanoic acid, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-22 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.368 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
2E5S
 
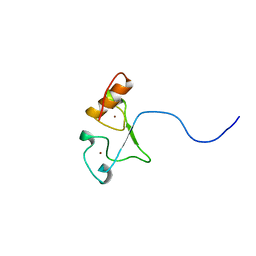 | | Solution structure of the zf-CCCHx2 domain of muscleblind-like 2, isoform 1 [Homo sapiens] | | Descriptor: | OTTHUMP00000018578, ZINC ION | | Authors: | Dang, W, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-12-22 | | Release date: | 2007-06-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RNA binding domain in the human muscleblind-like protein 2
Protein Sci., 18, 2009
|
|
6AAZ
 
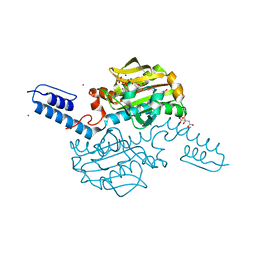 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with pNO2ZLys | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CITRIC ACID, MAGNESIUM ION, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-19 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.842 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
6AB8
 
 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with ZLys | | Descriptor: | (2S)-2-azanyl-6-(phenylmethoxycarbonylamino)hexanoic acid, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-20 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.753 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
6AB2
 
 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with oClZLys | | Descriptor: | (2S)-2-azanyl-6-[(2-chlorophenyl)methoxycarbonylamino]hexanoic acid, 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-19 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
6AAQ
 
 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with BCNLys | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, N6-({[(1R,8S,9s)-bicyclo[6.1.0]non-4-yn-9-yl]methoxy}carbonyl)-L-lysine, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-18 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
6AAN
 
 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with mEtZLys | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-18 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
2CU7
 
 | | Solution structure of the SANT domain of human KIAA1915 protein | | Descriptor: | KIAA1915 protein | | Authors: | Yoneyama, M, Umehara, T, Saito, K, Tochio, N, Koshiba, S, Inoue, M, Tanaka, A, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-25 | | Release date: | 2005-11-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Differences of SWIRM Domain Subtypes
J.Mol.Biol., 369, 2007
|
|
2COM
 
 | | The solution structure of the SWIRM domain of human LSD1 | | Descriptor: | Lysine-specific histone demethylase 1 | | Authors: | Tochio, N, Umehara, T, Koshiba, S, Inoue, M, Tanaka, A, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-18 | | Release date: | 2005-11-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SWIRM domain of human histone demethylase LSD1
Structure, 14, 2006
|
|
1UKX
 
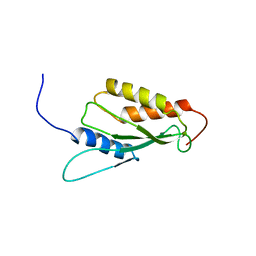 | | Solution structure of the RWD domain of mouse GCN2 | | Descriptor: | GCN2 eIF2alpha kinase | | Authors: | Nameki, N, Yoneyama, M, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-03 | | Release date: | 2004-08-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RWD domain of the mouse GCN2 protein.
Protein Sci., 13, 2004
|
|
1WMG
 
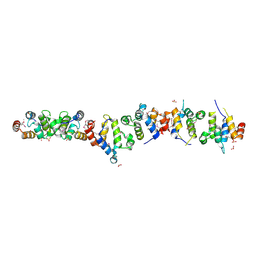 | | Crystal structure of the UNC5H2 death domain | | Descriptor: | SULFATE ION, SULFITE ION, netrin receptor Unc5h2 | | Authors: | Handa, N, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-07-09 | | Release date: | 2005-01-09 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the UNC5H2 death domain
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
1WWQ
 
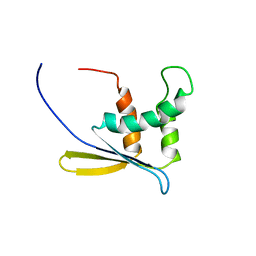 | | Solution Structure of Mouse ER | | Descriptor: | Enhancer of rudimentary homolog | | Authors: | Li, H, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-12 | | Release date: | 2006-01-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the mouse enhancer of rudimentary protein reveals a novel fold
J.Biomol.Nmr, 32, 2005
|
|
1UL7
 
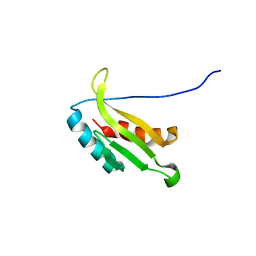 | | Solution structure of kinase associated domain 1 of mouse MAP/microtubule affinity-regulating kinase 3 | | Descriptor: | MAP/microtubule affinity-regulating kinase 3 | | Authors: | Tochio, N, Koshiba, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-10 | | Release date: | 2004-03-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the kinase-associated domain 1 of mouse microtubule-associated protein/microtubule affinity-regulating kinase 3
Protein Sci., 15, 2006
|
|
1X4S
 
 | | Solution structure of zinc finger HIT domain in protein FON | | Descriptor: | ZINC ION, Zinc finger HIT domain containing protein 2 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the zinc finger HIT domain in protein FON
Protein Sci., 16, 2007
|
|
1UGO
 
 | | Solution structure of the first Murine BAG domain of Bcl2-associated athanogene 5 | | Descriptor: | Bcl2-associated athanogene 5 | | Authors: | Endoh, H, Hayashi, F, Seimiya, K, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-17 | | Release date: | 2004-08-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The C-terminal BAG domain of BAG5 induces conformational changes of the Hsp70 nucleotide-binding domain for ADP-ATP exchange
Structure, 18, 2010
|
|
5FOR
 
 | | Cryptic TIR | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, PENTAETHYLENE GLYCOL, PHOSPHOINOSITIDE 3-KINASE ADAPTER PROTEIN 1 | | Authors: | Halabi, S, Gay, N.J, Moncrieffe, M.C. | | Deposit date: | 2015-11-25 | | Release date: | 2016-12-07 | | Last modified: | 2017-03-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Toll/Interleukin-1 Receptor (TIR) Domain of the B-cell Adaptor That Links Phosphoinositide Metabolism with the Negative Regulation of the Toll-like Receptor (TLR) Signalosome.
J. Biol. Chem., 292, 2017
|
|
